3KRO
 
 | | Mint heterotetrameric geranyl pyrophosphate synthase in complex with magnesium, IPP, and DMASPP (II) | | Descriptor: | 1,2-ETHANEDIOL, 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, DIMETHYLALLYL S-THIOLODIPHOSPHATE, ... | | Authors: | Chang, T.-H, Hsieh, F.-L, Ko, T.-P, Wang, A.H.-J. | | Deposit date: | 2009-11-19 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of a heterotetrameric geranyl pyrophosphate synthase from mint (Mentha piperita) reveals intersubunit regulation
Plant Cell, 22, 2010
|
|
9H5Q
 
 | | Crystal structure of Thermoanaerobacterales bacterium monoamine oxidase in complex with spermidine and its oxidation products | | Descriptor: | 1,3-DIAMINOPROPANE, 4-HYDROXYBUTAN-1-AMINIUM, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Basile, L, Poli, C, Santema, L.L, Lesenciuc, R.C, Fraaije, M.W, Binda, C. | | Deposit date: | 2024-10-23 | | Release date: | 2025-01-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Altering substrate specificity of a thermostable bacterial monoamine oxidase by structure-based mutagenesis.
Arch.Biochem.Biophys., 764, 2024
|
|
9HKW
 
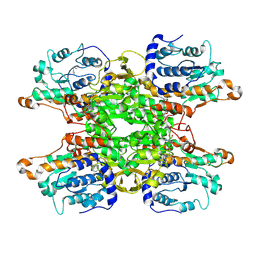 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 3 open and 1 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 3 open and 1 closed subunits
To Be Published
|
|
6UEH
 
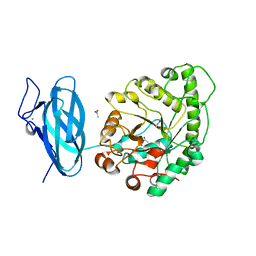 | | Crystal structure of a ruminal GH26 endo-beta-1,4-mannanase | | Descriptor: | ACETATE ION, CALCIUM ION, Cow rumen GH26 endo-mannanase | | Authors: | Mandelli, F, Morais, M.A.B, Lima, E.A, Persinoti, G.F, Murakami, M.T. | | Deposit date: | 2019-09-21 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.849 Å) | | Cite: | Spatially remote motifs cooperatively affect substrate preference of a ruminal GH26-type endo-beta-1,4-mannanase.
J.Biol.Chem., 295, 2020
|
|
9HKX
 
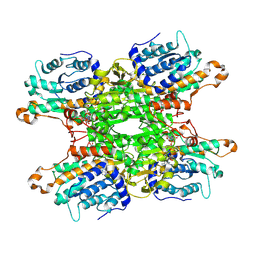 | | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 1 open and 3 closed subunits | | Descriptor: | ADENOSINE, Adenosylhomocysteinase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Malecki, P.H, Wozniak, K, Ruszkowski, M, Brzezinski, K. | | Deposit date: | 2024-12-04 | | Release date: | 2025-02-19 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Cryo-EM structure of Pseudomonas aeruginosa tetrameric S-adenosyl-L-homocysteine hydrolase with 1 open and 3 closed subunits
To Be Published
|
|
6UGT
 
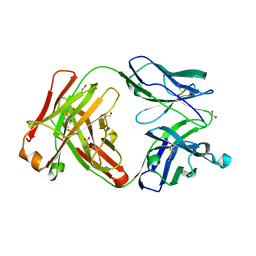 | | Crystal structure of the Fab fragment of PF06438179/GP1111 an infliximab biosimilar in a I-centered orthorhombic crystal form, Lot A | | Descriptor: | 1,2-ETHANEDIOL, PF06438179 Fab Heavy Chain, PF06438179 Fab Light Chain | | Authors: | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H.D. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
5GQK
 
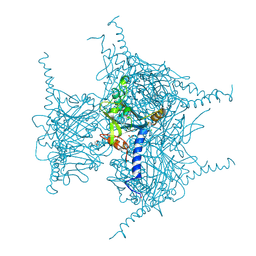 | | Crystal structure of Cypovirus Polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Polyhedrin | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
5GQN
 
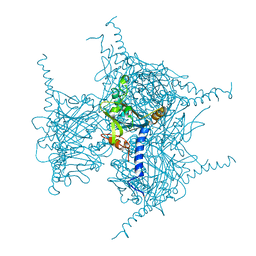 | | Crystal structure of in cellulo Cypovirus Polyhedra mutant with deletion of Gly192-Ala194 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Polyhedrin | | Authors: | Abe, S, Tabe, H, Ijiri, H, Yamashita, K, Hirata, K, Mori, H, Ueno, T. | | Deposit date: | 2016-08-07 | | Release date: | 2017-02-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Engineering of Self-Assembled Porous Protein Materials in Living Cells
ACS Nano, 11, 2017
|
|
6UGV
 
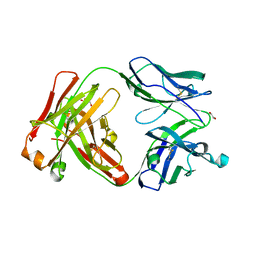 | | Crystal structure of the Fab fragment of anti-TNFa antibody infliximab (Remicade) in a I-centered orthorhombic crystal form, Lot C | | Descriptor: | 1,2-ETHANEDIOL, Infliximab Fab Heavy Chain, Infliximab Fab Light Chain, ... | | Authors: | Lerch, T.F, Sharpe, P, Mayclin, S.J, Edwards, T.E, Polleck, S, Rouse, J.C, Conlan, H.D. | | Deposit date: | 2019-09-26 | | Release date: | 2019-11-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of PF-06438179/GP1111, an Infliximab Biosimilar.
BioDrugs, 34, 2020
|
|
5LOM
 
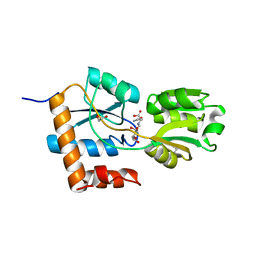 | | Crystal structure of the PBP SocA from Agrobacterium tumefaciens C58 in complex with DFG at 1.5 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Deoxyfructosyl-amino Acid Transporter Periplasmic Binding Protein, Deoxyfructosylglutamine | | Authors: | Marty, L, Vigouroux, A, Morera, S. | | Deposit date: | 2016-08-09 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Basis for High Specificity of Amadori Compound and Mannopine Opine Binding in Bacterial Pathogens.
J.Biol.Chem., 291, 2016
|
|
6UFW
 
 | |
3MBA
 
 | | APLYSIA LIMACINA MYOGLOBIN. CRYSTALLOGRAPHIC ANALYSIS AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | FLUORIDE ION, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bolognesi, M, Onesti, S, Gatti, G, Coda, A, Ascenzi, P, Brunori, M. | | Deposit date: | 1989-02-22 | | Release date: | 1990-01-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Aplysia limacina myoglobin. Crystallographic analysis at 1.6 A resolution.
J.Mol.Biol., 205, 1989
|
|
5IS7
 
 | | Crystal structure of mouse CARM1 in complex with decarboxylated SAH | | Descriptor: | 1,2-ETHANEDIOL, 5'-S-(3-aminopropyl)-5'-thioadenosine, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-03-15 | | Release date: | 2017-03-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with decarboxylated SAH
To Be Published
|
|
5CLH
 
 | |
6CAD
 
 | | Crystal structure of RAF kinase domain bound to the inhibitor 2a | | Descriptor: | 1-(propan-2-yl)-3-({3-[3-(trifluoromethyl)phenyl]isoquinolin-8-yl}ethynyl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Serine/threonine-protein kinase B-raf | | Authors: | Maisonneuve, P, Kurinov, I, Assadieskandar, A, Yu, C, Liu, X, Chen, Y.-C, Prakash, G.K.S, Zhang, C, SIcheri, F. | | Deposit date: | 2018-01-30 | | Release date: | 2018-02-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Effects of rigidity on the selectivity of protein kinase inhibitors.
Eur J Med Chem, 146, 2018
|
|
6CC2
 
 | | Crystal Structure of CDC45 from Entamoeba histolytica | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cell division control protein 45 cdc45 putative, ... | | Authors: | Shi, K, Kurniawan, F, Kurahashi, K, Bielinsky, A, Aihara, H. | | Deposit date: | 2018-02-05 | | Release date: | 2018-06-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal Structure ofEntamoeba histolyticaCdc45 Suggests a Conformational Switch that May Regulate DNA Replication.
iScience, 3, 2018
|
|
1P3X
 
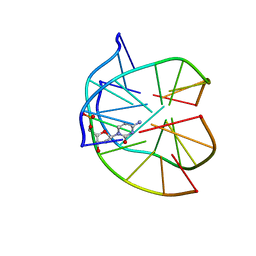 | | INTRAMOLECULAR DNA TRIPLEX WITH 1-PROPYNYL DEOXYURIDINE IN THE THIRD STRAND, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*(PDU)P*CP*(PDU)P*(DCM)P*(PDU)P*CP*(PDU)P*(PDU))-3'), DNA (5'-D(*AP*GP*AP*GP*AP*GP*AP*A)-3'), DNA (5'-D(*TP*TP*CP*TP*CP*TP*CP*T)-3') | | Authors: | Phipps, A.K, Tarkoy, M, Schultze, P, Feigon, J. | | Deposit date: | 1998-02-05 | | Release date: | 1998-05-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an intramolecular DNA triplex containing 5-(1-propynyl)-2'-deoxyuridine residues in the third strand.
Biochemistry, 37, 1998
|
|
1Y66
 
 | | Dioxane contributes to the altered conformation and oligomerization state of a designed engrailed homeodomain variant | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, ACETIC ACID, CADMIUM ION, ... | | Authors: | Hom, G.K, Lassila, J.K, Thomas, L.M, Mayo, S.L. | | Deposit date: | 2004-12-03 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Dioxane contributes to the altered conformation and oligomerization state of a designed engrailed homeodomain variant.
Protein Sci., 14, 2005
|
|
6S4D
 
 | |
6S51
 
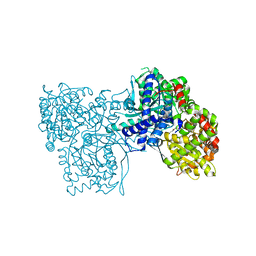 | | The crystal structure of glycogen phosphorylase in complex with 10 | | Descriptor: | (2~{R},3~{S},4~{R},5~{R},6~{S})-2-(hydroxymethyl)-6-(2-phenyl-1,3-thiazol-4-yl)oxane-3,4,5-triol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Solovou, T.G.A, Papaioannou, O.S.E, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2019-06-29 | | Release date: | 2020-02-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | The architecture of hydrogen and sulfur sigma-hole interactions explain differences in the inhibitory potency of C-beta-d-glucopyranosyl thiazoles, imidazoles and an N-beta-d glucopyranosyl tetrazole for human liver glycogen phosphorylase and offer new insights to structure-based design.
Bioorg.Med.Chem., 28, 2020
|
|
6ZYP
 
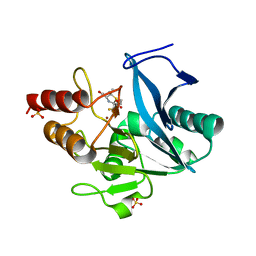 | | Structure of NDM-1 with 2-Mercaptomethyl-thiazolidine L-anti-1b | | Descriptor: | (2~{S},4~{R})-2-ethoxycarbonyl-5,5-dimethyl-2-(sulfanylmethyl)-1,3-thiazolidine-4-carboxylic acid, Metallo-beta-lactamase type 2, SULFATE ION, ... | | Authors: | Hinchliffe, P, Spencer, J. | | Deposit date: | 2020-08-02 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | 2-Mercaptomethyl-thiazolidines use conserved aromatic-S interactions to achieve broad-range inhibition of metallo-beta-lactamases.
Chem Sci, 12, 2021
|
|
5FWA
 
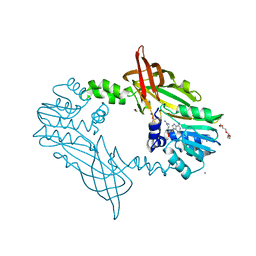 | | Crystal Structure of Mus musculus Protein Arginine Methyltransferase 2 with CP1 | | Descriptor: | 1,2-ETHANEDIOL, 9-(6-carbamimidamido-5,6-dideoxy-beta-D-ribo-hexofuranosyl)-9H-purin-6-amine, CALCIUM ION, ... | | Authors: | Cura, V, Troffer-Charlier, N, Marechal, N, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-02-16 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural studies of protein arginine methyltransferase 2 reveal its interactions with potential substrates and inhibitors.
FEBS J., 284, 2017
|
|
8E73
 
 | | Vigna radiata supercomplex I+III2 (full bridge) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, CARDIOLIPIN, ... | | Authors: | Maldonado, M, Letts, J.A. | | Deposit date: | 2022-08-23 | | Release date: | 2023-01-11 | | Last modified: | 2024-12-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Plant-specific features of respiratory supercomplex I + III 2 from Vigna radiata.
Nat.Plants, 9, 2023
|
|
7Q45
 
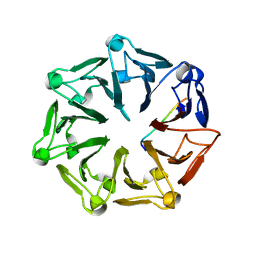 | | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of Myelin transcription factor 1 | | Descriptor: | CITRIC ACID, E3 ubiquitin-protein ligase HERC2, Myelin transcription factor 1 | | Authors: | Demenge, A, Howard, E, Cousido-Siah, A, Mitschler, A, Podjarny, A, McEwen, A.G, Trave, G. | | Deposit date: | 2021-10-29 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.09999585 Å) | | Cite: | Crystal structure of RCC1-Like domain 2 of ubiquitin ligase HERC2 in complex with DXDKDED motif of Myelin transcription factor 1
To Be Published
|
|
4Y9X
 
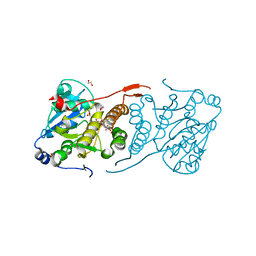 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis in complex with Mn2+, uridine-diphosphate-glucose (UDP-Glc) and phosphoglyceric acid (PGA) - GpgS Mn2+ UDP-Glc PGA-3 | | Descriptor: | 1,2-ETHANEDIOL, 3-PHOSPHOGLYCERIC ACID, CHLORIDE ION, ... | | Authors: | Albesa-Jove, D, Rodrigo-Unzueta, A, Cifuente, J.O, Urresti, S, Comino, N, Sancho-Vaello, E, Guerin, M.E. | | Deposit date: | 2015-02-17 | | Release date: | 2015-07-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.637 Å) | | Cite: | A Native Ternary Complex Trapped in a Crystal Reveals the Catalytic Mechanism of a Retaining Glycosyltransferase.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
