1HZU
 
 | | DOMAIN SWING UPON HIS TO ALA MUTATION IN NITRITE REDUCTASE OF PSEUDOMONAS AERUGINOSA | | Descriptor: | HEME C, HEME D, NITRITE REDUCTASE | | Authors: | Brown, K, Cutruzzola, F, Brunori, M, Tegoni, M, Cambillau, C. | | Deposit date: | 2001-01-26 | | Release date: | 2001-09-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Domain swing upon His to Ala mutation in nitrite reductase of Pseudomonas aeruginosa.
J.Mol.Biol., 312, 2001
|
|
2WCX
 
 | |
2G00
 
 | | Factor Xa in complex with the inhibitor 3-(6-(2'-((dimethylamino)methyl)-4-biphenylyl)-7-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-c]pyridin-1-yl)benzamide | | Descriptor: | 3-[6-{2'-[(DIMETHYLAMINO)METHYL]BIPHENYL-4-YL}-7-OXO-3-(TRIFLUOROMETHYL)-4,5,6,7-TETRAHYDRO-1H-PYRAZOLO[3,4-C]PYRIDIN-1-YL]BENZAMIDE, Coagulation factor X | | Authors: | Alexander, R.S. | | Deposit date: | 2006-02-10 | | Release date: | 2006-10-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of potent, efficacious, and orally bioavailable inhibitors of blood coagulation factor Xa with neutral P1 moieties.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4JB4
 
 | | Expression, Purification, Characterization, and Solution NMR Study of Highly Deuterated Yeast Cytochrome c Peroxidase with Enhanced Solubility | | Descriptor: | Cytochrome c peroxidase, mitochondrial, FLUORIDE ION, ... | | Authors: | Wohlkonig, A.C. | | Deposit date: | 2013-02-19 | | Release date: | 2013-04-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Expression, purification, characterization, and solution nuclear magnetic resonance study of highly deuterated yeast cytochrome C peroxidase with enhanced solubility.
Biochemistry, 52, 2013
|
|
2B1G
 
 | | Crystal structures of transition state analogue inhibitors of inosine monophosphate cyclohydrolase | | Descriptor: | 7-(3,4-DIHYDROXY-5R-HYDROXYMETHYLTETRAHYDROFURAN-2-YL)-2,2-DIOXO-1,2R,3R,7-TETRAHYDRO-2L6-IMIDAZO[4,5-C][1,2,6]THIADIAZIN-4S-ONE, Bifunctional purine biosynthesis protein PURH, PHOSPHATE ION, ... | | Authors: | Xu, L, Chong, Y, Hwang, I, D'Onofrio, A, Amore, K, Beardsley, G.P, Li, C, Olson, A.J, Boger, D.L, Wilson, I.A. | | Deposit date: | 2005-09-15 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based Design, Synthesis, Evaluation, and Crystal Structures of Transition State Analogue Inhibitors of Inosine Monophosphate Cyclohydrolase.
J.Biol.Chem., 282, 2007
|
|
4JXT
 
 | | CID of human RPRD1A in complex with a phosphorylated peptide from RPB1-CTD | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, Regulation of nuclear pre-mRNA domain-containing protein 1A, UNKNOWN ATOM OR ION | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-28 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
2JJL
 
 | | Structure of avian reovirus sigmaC117-326, P321 crystal form | | Descriptor: | CHLORIDE ION, SIGMA-C CAPSID PROTEIN, SULFATE ION, ... | | Authors: | Guardado-Calvo, P, Fox, G.C, Llamas-Saiz, A.L, Benavente, J, van Raaij, M.J. | | Deposit date: | 2008-04-14 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic structure of the alpha-helical triple coiled-coil domain of avian reovirus S1133 fibre.
J. Gen. Virol., 90, 2009
|
|
3NKX
 
 | |
2ORL
 
 | | Solution structure of the cytochrome c- para-aminophenol adduct | | Descriptor: | 4-AMINOPHENOL, Cytochrome c iso-1, HEME C | | Authors: | Assfalg, M, Bertini, I, Del Conte, R, Giachetti, A, Turano, P. | | Deposit date: | 2007-02-03 | | Release date: | 2007-04-24 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Cytochrome c and organic molecules: solution structure of the p-aminophenol adduct.
Biochemistry, 46, 2007
|
|
1YRZ
 
 | | Crystal structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125 | | Descriptor: | xylan beta-1,4-xylosidase | | Authors: | Fedorov, A.A, Fedorov, E.V, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-02-05 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of xylan beta-1,4-xylosidase from Bacillus Halodurans C-125
To be Published
|
|
3PLF
 
 | | Reverse Binding Mode of MetRD peptide complexed with c-Cbl TKB domain | | Descriptor: | CALCIUM ION, CHLORIDE ION, E3 ubiquitin-protein ligase CBL, ... | | Authors: | Sun, Q, Sivaraman, J. | | Deposit date: | 2010-11-15 | | Release date: | 2010-12-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | An adjacent arginine, and the phosphorylated tyrosine in the c-Met receptor target sequence, dictates the orientation of c-Cbl binding
Febs Lett., 585, 2011
|
|
1E8E
 
 | | Solution Structure of Methylophilus methylotrophus Cytochrome c''. Insights into the Structural Basis of Haem-Ligand Detachment | | Descriptor: | CYTOCHROME C'', HEME C | | Authors: | Brennan, L, Turner, D.L, Fareleira, P, Santos, H. | | Deposit date: | 2000-09-20 | | Release date: | 2001-09-20 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Methylophilus Methylotrophus Cytochrome C": Insights Into the Structural Basis of Haem-Ligand Detachment
J.Mol.Biol., 308, 2001
|
|
3U6H
 
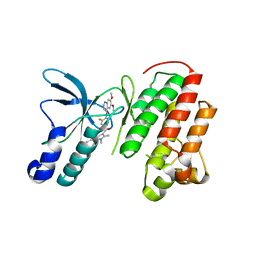 | | Crystal structure of c-Met in complex with pyrazolone inhibitor 26 | | Descriptor: | Hepatocyte growth factor receptor, N-{4-[(6,7-dimethoxyquinolin-4-yl)oxy]-3-fluorophenyl}-1,5-dimethyl-3-oxo-2-phenyl-2,3-dihydro-1H-pyrazole-4-carboxamide | | Authors: | Bellon, S.F, Whittington, D.A, Long, A.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of novel class II c-Met inhibitors: 1. Identification of pyrazolone-based derivatives.
J.Med.Chem., 55, 2012
|
|
1FS7
 
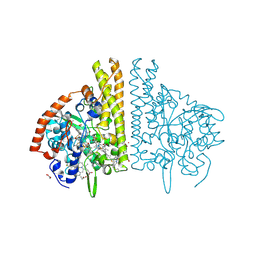 | | CYTOCHROME C NITRITE REDUCTASE FROM WOLINELLA SUCCINOGENES | | Descriptor: | ACETATE ION, CALCIUM ION, CYTOCHROME C NITRITE REDUCTASE, ... | | Authors: | Einsle, O, Stach, P, Messerschmidt, A, Simon, J, Kroeger, A, Huber, R, Kroneck, P.M.H. | | Deposit date: | 2000-09-08 | | Release date: | 2001-01-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cytochrome c nitrite reductase from Wolinella succinogenes. Structure at 1.6 A resolution, inhibitor binding, and heme-packing motifs.
J.Biol.Chem., 275, 2000
|
|
1FIC
 
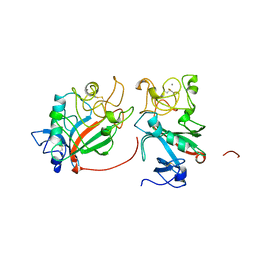 | |
2LGW
 
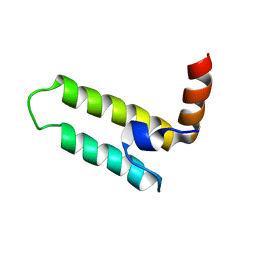 | | Solution Structure of the J Domain of HSJ1a | | Descriptor: | DnaJ homolog subfamily B member 2 | | Authors: | Zhou, C, Gao, X, Cao, C, Hu, H. | | Deposit date: | 2011-08-02 | | Release date: | 2012-01-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-terminal helices of heat shock protein 70 are essential for J-domain binding and ATPase activation.
J.Biol.Chem., 287, 2012
|
|
2HE2
 
 | | Crystal structure of the 3rd PDZ domain of human discs large homologue 2, DLG2 | | Descriptor: | Discs large homolog 2 | | Authors: | Turnbull, A.P, Phillips, C, Berridge, G, Savitsky, P, Smee, C.E.A, Papagrigoriou, E, Debreczeni, J, Gorrec, F, Elkins, J.M, von Delft, F, Weigelt, J, Edwards, A, Arrowsmith, C, Sundstrom, M, Doyle, D.A, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of PICK1 and other PDZ domains obtained with the help of self-binding C-terminal extensions.
Protein Sci., 16, 2007
|
|
1U75
 
 | |
1LQB
 
 | | Crystal structure of a hydroxylated HIF-1 alpha peptide bound to the pVHL/elongin-C/elongin-B complex | | Descriptor: | Elongin B, Elongin C, Hypoxia-inducible factor 1 ALPHA, ... | | Authors: | Hon, W.C, Wilson, M.I, Harlos, K, Claridge, T.D, Schofield, C.J, Pugh, C.W, Maxwell, P.H, Ratcliffe, P.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2002-05-09 | | Release date: | 2002-07-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the recognition of hydroxyproline in HIF-1 alpha by pVHL.
Nature, 417, 2002
|
|
3R7O
 
 | | Structure of dually phosphorylated c-MET receptor kinase in complex with an MK-2461 analog | | Descriptor: | Hepatocyte growth factor receptor, N-[(2R)-1,4-dioxan-2-ylmethyl]-N-methyl-N'-{5-oxo-3-[1-(piperidin-4-yl)-1H-pyrazol-4-yl]-5H-benzo[4,5]cyclohepta[1,2-b]pyridin-7-yl}sulfuric diamide | | Authors: | Soisson, S.M, Rickert, K, Patel, S.B, Munshi, S, Lumb, K.J. | | Deposit date: | 2011-03-22 | | Release date: | 2012-02-01 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for selective small molecule kinase inhibition of activated c-Met.
J.Biol.Chem., 286, 2011
|
|
1CYC
 
 | | THE CRYSTAL STRUCTURE OF BONITO (KATSUO) FERROCYTOCHROME C AT 2.3 ANGSTROMS RESOLUTION. II. STRUCTURE AND FUNCTION | | Descriptor: | FERROCYTOCHROME C, HEME C | | Authors: | Tanaka, N, Yamane, T, Tsukihara, T, Ashida, T, Kakudo, M. | | Deposit date: | 1976-08-01 | | Release date: | 1976-10-06 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of bonito (katsuo) ferrocytochrome c at 2.3 A resolution. II. Structure and function.
J.Biochem.(Tokyo), 77, 1975
|
|
1ZTG
 
 | | human alpha polyC binding protein KH1 | | Descriptor: | 5'-D(P*CP*CP*CP*TP*CP*CP*CP*T)-3', POLY(RC)-BINDING PROTEIN 1 | | Authors: | Sidiqi, M, Wilce, J.A, Barker, A, Schmidgerger, J, Leedman, P.J, Wilce, M.C.J. | | Deposit date: | 2005-05-27 | | Release date: | 2006-05-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Contribution of the first K-homology domain of poly(C)-binding protein 1 to its affinity and specificity for C-rich oligonucleotides
Nucleic Acids Res., 40, 2012
|
|
1CRC
 
 | | CYTOCHROME C AT LOW IONIC STRENGTH | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Sanishvili, R, Volz, K.W, Westbrook, E.M, Margoliash, E. | | Deposit date: | 1995-03-22 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The low ionic strength crystal structure of horse cytochrome c at 2.1 A resolution and comparison with its high ionic strength counterpart.
Structure, 3, 1995
|
|
1CI3
 
 | | CYTOCHROME F FROM THE B6F COMPLEX OF PHORMIDIUM LAMINOSUM | | Descriptor: | HEME C, PROTEIN (CYTOCHROME F), ZINC ION | | Authors: | Carrell, C.J, Schlarb, B.G, Howe, C.J, Bendall, D.S, Cramer, W.A, Smith, J.L. | | Deposit date: | 1999-04-07 | | Release date: | 1999-08-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the soluble domain of cytochrome f from the cyanobacterium Phormidium laminosum.
Biochemistry, 38, 1999
|
|
2IWN
 
 | | 3rd PDZ domain of Multiple PDZ Domain Protein MPDZ | | Descriptor: | MULTIPLE PDZ DOMAIN PROTEIN | | Authors: | Elkins, J.M, Gileadi, C, Savitsky, P, Berridge, G, Smee, C.E.A, Pike, A.C.W, Papagrigoriou, E, Sundstrom, M, Edwards, A, Arrowsmith, C, Weigelt, J, Doyle, D.A. | | Deposit date: | 2006-07-03 | | Release date: | 2006-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of Pick1 and Other Pdz Domains Obtained with the Help of Self-Binding C-Terminal Extension.
Protein Sci., 16, 2007
|
|
