1NJK
 
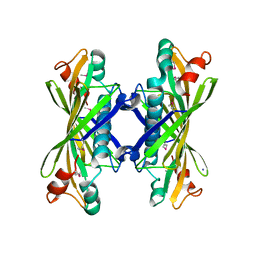 | | Crystal Structure of YbaW Probable Thioesterase from Escherichia coli | | Descriptor: | Hypothetical protein ybaW, IODIDE ION | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-31 | | Release date: | 2003-07-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Escherichia coli Hypothetical Protein YbaW
To be Published
|
|
1NJZ
 
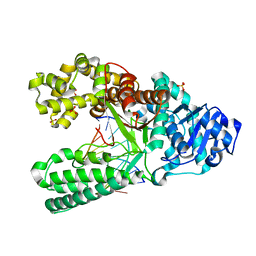 | |
1NK9
 
 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER TWO ROUNDS OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP AND DGTP. | | Descriptor: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
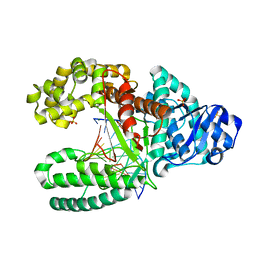
1NJ1
 
 | | Crystal structure of Prolyl-tRNA Synthetase from Methanothermobacter thermautotrophicus bound to cysteine sulfamoyl adenylate | | Descriptor: | 5'-O-(N-(L-CYSTEINYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, Proline-tRNA Synthetase, ... | | Authors: | Kamtekar, S, Kennedy, W.D, Wang, J, Stathopoulos, C, Soll, D, Steitz, T.A. | | Deposit date: | 2002-12-30 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structural basis of cysteine aminoacylation of tRNAPro by prolyl-tRNA synthetases
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NL1
 
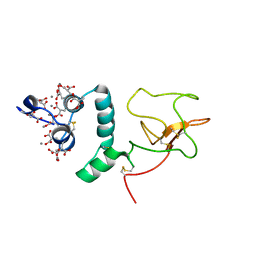 | | BOVINE PROTHROMBIN FRAGMENT 1 IN COMPLEX WITH CALCIUM ION | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Huang, M, Huang, G, Furie, B, Seaton, B, Furie, B.C. | | Deposit date: | 2003-01-06 | | Release date: | 2003-09-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of membrane binding by Gla domains of vitamin K-dependent proteins.
Nat.Struct.Biol., 10, 2003
|
|
4F3Z
 
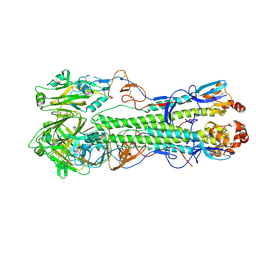 | | Crystal structure of a swine H1N2 influenza virus hemagglutinin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Xu, R, Wilson, I.A. | | Deposit date: | 2012-05-09 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Functional Balance of the Hemagglutinin and Neuraminidase Activities Accompanies the Emergence of the 2009 H1N1 Influenza Pandemic.
J.Virol., 86, 2012
|
|
1F1T
 
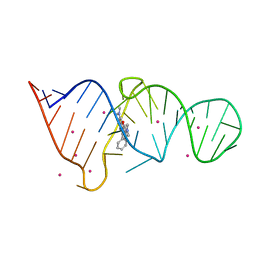 | | CRYSTAL STRUCTURE OF THE MALACHITE GREEN APTAMER COMPLEXED WITH TETRAMETHYL-ROSAMINE | | Descriptor: | MALACHITE GREEN APTAMER RNA, N,N'-TETRAMETHYL-ROSAMINE, STRONTIUM ION | | Authors: | Baugh, C, Grate, D, Wilson, C. | | Deposit date: | 2000-05-19 | | Release date: | 2000-09-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 2.8 A crystal structure of the malachite green aptamer.
J.Mol.Biol., 301, 2000
|
|
1NK1
 
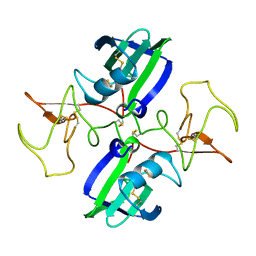 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR/SCATTER FACTOR (HGF/SF) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (HEPATOCYTE GROWTH FACTOR PRECURSOR) | | Authors: | Chirgadze, D.Y, Hepple, J.P, Zhou, H, Byrd, R.A, Blundell, T.L, Gherardi, E. | | Deposit date: | 1998-08-20 | | Release date: | 1999-01-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Nat.Struct.Biol., 6, 1999
|
|
1NGD
 
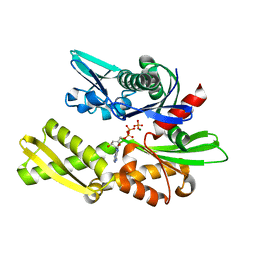 | | STRUCTURAL BASIS OF THE 70-KILODALTON HEAT SHOCK COGNATE PROTEIN ATP HYDROLYTIC ACTIVITY, II. STRUCTURE OF THE ACTIVE SITE WITH ADP OR ATP BOUND TO WILD TYPE AND MUTANT ATPASE FRAGMENT | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HEAT-SHOCK COGNATE 70 kD PROTEIN, MAGNESIUM ION, ... | | Authors: | Flaherty, K.M, Wilbanks, S.M, Deluca-Flaherty, C, Mckay, D.B. | | Deposit date: | 1994-05-17 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis of the 70-kilodalton heat shock cognate protein ATP hydrolytic activity. II. Structure of the active site with ADP or ATP bound to wild type and mutant ATPase fragment.
J.Biol.Chem., 269, 1994
|
|
1NK8
 
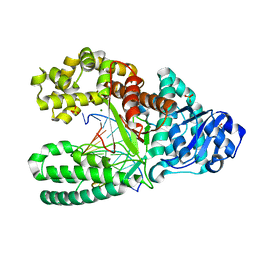 | | A BACILLUS DNA POLYMERASE I PRODUCT COMPLEX BOUND TO A GUANINE-THYMINE MISMATCH AFTER A SINGLE ROUND OF PRIMER EXTENSION, FOLLOWING INCORPORATION OF DCTP. | | Descriptor: | DNA POLYMERASE I, DNA PRIMER STRAND, DNA TEMPLATE STRAND, ... | | Authors: | Johnson, S.J, Beese, L.S. | | Deposit date: | 2003-01-02 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structures of mismatch replication errors observed in a DNA polymerase.
Cell(Cambridge,Mass.), 116, 2004
|
|
1NE2
 
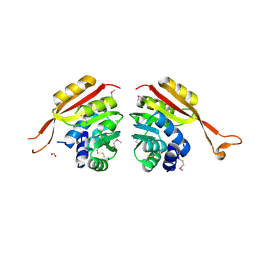 | | Crystal Structure of Thermoplasma acidophilum 1320 (APC5513) | | Descriptor: | FORMIC ACID, hypothetical protein ta1320 | | Authors: | Kim, Y, Joachimiak, A, Edwards, A, Xu, X, Christendat, D, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-12-10 | | Release date: | 2003-07-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Thermoplasma acidophilum 1320 (APC5513)
To be Published
|
|
1NFD
 
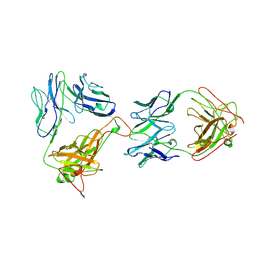 | | AN ALPHA-BETA T CELL RECEPTOR (TCR) HETERODIMER IN COMPLEX WITH AN ANTI-TCR FAB FRAGMENT DERIVED FROM A MITOGENIC ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, H57 FAB, N15 ALPHA-BETA T-CELL RECEPTOR | | Authors: | Wang, J.-H, Lim, K, Smolyar, A, Teng, M.-K, Sacchittini, J, Reinherz, E.L. | | Deposit date: | 1997-08-04 | | Release date: | 1998-01-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Atomic structure of an alphabeta T cell receptor (TCR) heterodimer in complex with an anti-TCR fab fragment derived from a mitogenic antibody.
EMBO J., 17, 1998
|
|
1NH2
 
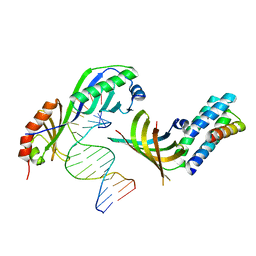 | | Crystal structure of a yeast TFIIA/TBP/DNA complex | | Descriptor: | 5'-D(*GP*TP*TP*TP*TP*AP*TP*AP*TP*AP*CP*AP*TP*AP*CP*A)-3', 5'-D(*TP*GP*TP*AP*(5IU)P*GP*TP*AP*TP*AP*(5IU)P*AP*AP*AP*AP*C)-3', Transcription initiation factor IIA large chain, ... | | Authors: | Bleichenbacher, M, Tan, S, Richmond, T.J. | | Deposit date: | 2002-12-18 | | Release date: | 2003-10-21 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Novel interactions between the components of human and yeast TFIIA/TBP/DNA complexes.
J.Mol.Biol., 332, 2003
|
|
1PRX
 
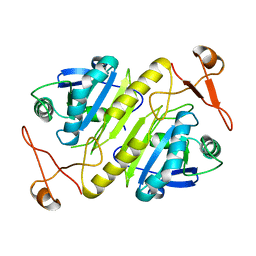 | | HORF6 A NOVEL HUMAN PEROXIDASE ENZYME | | Descriptor: | HORF6 | | Authors: | Choi, H.-J, Kang, S.W, Yang, C.-H, Rhee, S.G, Ryu, S.-E. | | Deposit date: | 1998-04-03 | | Release date: | 1998-06-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a novel human peroxidase enzyme at 2.0 A resolution.
Nat.Struct.Biol., 5, 1998
|
|
1PME
 
 | |
1PTU
 
 | |
1PTY
 
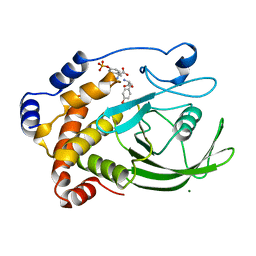 | | CRYSTAL STRUCTURE OF PROTEIN TYROSINE PHOSPHATASE 1B COMPLEXED WITH TWO PHOSPHOTYROSINE MOLECULES | | Descriptor: | MAGNESIUM ION, O-PHOSPHOTYROSINE, PROTEIN TYROSINE PHOSPHATASE 1B | | Authors: | Zhao, Y, Puius, Y.A, Sullivan, M, Lawrence, D, Almo, S.C, Zhang, Z.-Y. | | Deposit date: | 1997-01-16 | | Release date: | 1998-01-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Identification of a second aryl phosphate-binding site in protein-tyrosine phosphatase 1B: a paradigm for inhibitor design.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
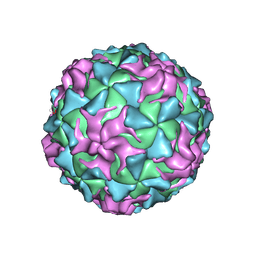
1POV
 
 | |
1PHS
 
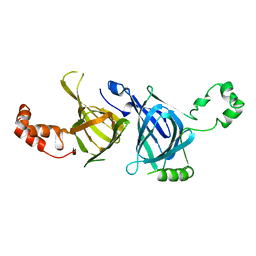 | | THE THREE-DIMENSIONAL STRUCTURE OF THE SEED STORAGE PROTEIN PHASEOLIN AT 3 ANGSTROMS RESOLUTION | | Descriptor: | PHASEOLIN, BETA-TYPE PRECURSOR | | Authors: | Lawrence, M.C, Suzuki, E, Varghese, J.N, Davis, P.C, Vandonkelaar, A, Tulloch, P.A, Colman, P.M. | | Deposit date: | 1990-03-21 | | Release date: | 1990-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The three-dimensional structure of the seed storage protein phaseolin at 3 A resolution.
EMBO J., 9, 1990
|
|
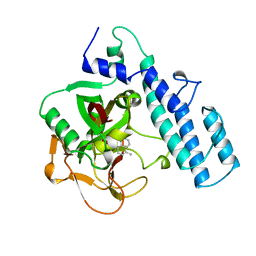
1PAX
 
 | |
1PBN
 
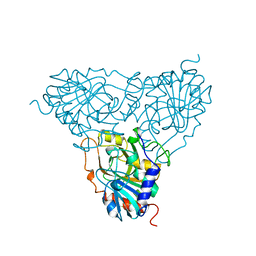 | | PURINE NUCLEOSIDE PHOSPHORYLASE | | Descriptor: | PURINE NUCLEOSIDE PHOSPHORYLASE | | Authors: | Mao, C, Ealick, S.E. | | Deposit date: | 1995-07-10 | | Release date: | 1995-11-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calf spleen purine nucleoside phosphorylase complexed with substrates and substrate analogues.
Biochemistry, 37, 1998
|
|
1PBG
 
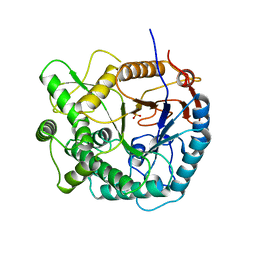 | |
1PAH
 
 | |
1PBK
 
 | | HOMOLOGOUS DOMAIN OF HUMAN FKBP25 | | Descriptor: | FKBP25, RAPAMYCIN IMMUNOSUPPRESSANT DRUG | | Authors: | Liang, J, Hung, D.T, Schreiber, S.L, Clardy, J. | | Deposit date: | 1995-09-01 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the human 25 kDa FK506 binding protein complexed with rapamycin.
J.Am.Chem.Soc., 118, 1996
|
|
1PHT
 
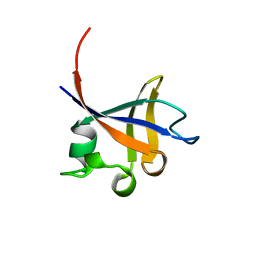 | | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT SH3 DOMAIN, RESIDUES 1-85 | | Descriptor: | PHOSPHATIDYLINOSITOL 3-KINASE P85-ALPHA SUBUNIT | | Authors: | Liang, J, Chen, J.K, Schreiber, S.L, Clardy, J. | | Deposit date: | 1995-08-17 | | Release date: | 1995-12-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of P13K SH3 domain at 20 angstroms resolution.
J.Mol.Biol., 257, 1996
|
|
