8P1T
 
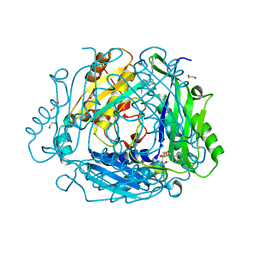 | | Crystal structure of human methionine adenosyltransferase 2A (MAT2A) in complex with SAM and allosteric inhibitor Z237451470 | | Descriptor: | 1,2-ETHANEDIOL, 6-cyclopropyl-~{N}-(1~{H}-indazol-5-yl)-1-propan-2-yl-pyrazolo[3,4-b]pyridine-4-carboxamide, CHLORIDE ION, ... | | Authors: | Thomsen, M, Thieulin-Pardo, G, Neumann, L. | | Deposit date: | 2023-05-12 | | Release date: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.442 Å) | | Cite: | Discovery of novel methionine adenosyltransferase 2A (MAT2A) allosteric inhibitors by structure-based virtual screening.
Bioorg.Med.Chem.Lett., 94, 2023
|
|
8P1S
 
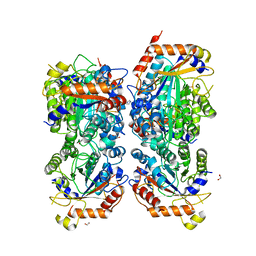 | | Bifidobacterium asteroides alpha-L-fucosidase (TT1819) in complex with fucose. | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifidobacterium asteroides alpha-L-fucosidase (TT1819), ... | | Authors: | Owen, C.D, Penner, M, Gascuena, A.N, Wu, H, Hernando, P.J, Monaco, S, Le Gall, G, Gardner, R, Ndeh, D, Urbanowicz, P.A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exploring sequence, structure and function of microbial fucosidases from glycoside hydrolase GH29 family
To Be Published
|
|
8P1R
 
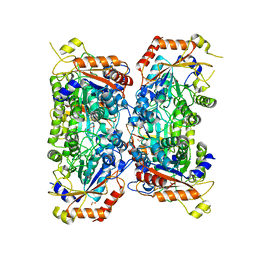 | | Bifidobacterium asteroides alpha-L-fucosidase (TT1819) catalytic mutant. | | Descriptor: | 1,2-ETHANEDIOL, Bifidobacterium asteroides alpha-L-fucosidase (TT1819) catalytic mutant, MAGNESIUM ION | | Authors: | Owen, C.D, Penner, M, Gascuena, A.N, Wu, H, Hernando, P.J, Monaco, S, Le Gall, G, Gardner, R, Ndeh, D, Urbanowicz, P.A, Spencer, D.I.R, Walsh, M.A, Angulo, J, Juge, N. | | Deposit date: | 2023-05-12 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Exploring sequence, structure and function of microbial fucosidases from glycoside hydrolase GH29 family
To Be Published
|
|
8P1O
 
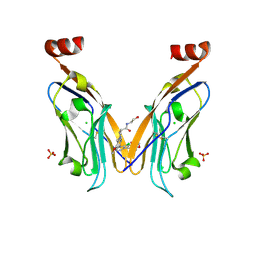 | | Solubilizer tag effect on PD-L1/inhibitor binding properties for m-terphenyl derivatives | | Descriptor: | (3~{R})-1-[[4-[2-chloranyl-3-(2,3-dihydro-1,4-benzodioxin-6-yl)phenyl]-2-methoxy-phenyl]methyl]-~{N}-(2-hydroxyethyl)pyrrolidine-3-carboxamide, CHLORIDE ION, Programmed cell death 1 ligand 1, ... | | Authors: | Plewka, J, Magiera-Mularz, K, Surmiak, E, Kalinowska-Tluscik, J, Holak, T.A. | | Deposit date: | 2023-05-12 | | Release date: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Solubilizer Tag Effect on PD-L1/Inhibitor Binding Properties for m -Terphenyl Derivatives.
Acs Med.Chem.Lett., 15, 2024
|
|
8P1I
 
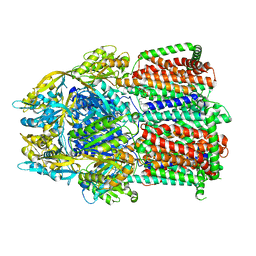 | | Single particle cryo-EM co-structure of Klebsiella pneumoniae AcrB with the BDM91288 efflux pump inhibitor at 2.97 Angstrom resolution | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-[PHOSPHO-RAC-(1-GLYCEROL), 3-chloranyl-2,6-di(piperazin-4-ium-1-yl)quinoline, Efflux pump membrane transporter, ... | | Authors: | Boernsen, C, Mueller, R.T, Pos, K.M, Frangakis, A.S. | | Deposit date: | 2023-05-12 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-31 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Pyridylpiperazine efflux pump inhibitor boosts in vivo antibiotic efficacy against K. pneumoniae.
Embo Mol Med, 16, 2024
|
|
8P1G
 
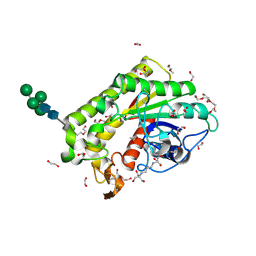 | | Crystal structure of inactive TtCE16 in complex with acetate | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Dimarogona, M, Kosinas, C, Pentari, C, Zerva, A, Topakas, E, Karampa, P. | | Deposit date: | 2023-05-12 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The role of CE16 exo-deacetylases in hemicellulolytic enzyme mixtures revealed by the biochemical and structural study of the novel TtCE16B esterase.
Carbohydr Polym, 327, 2024
|
|
8P1E
 
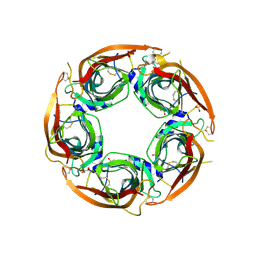 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL001613. | | Descriptor: | 1-[4-(trifluoromethyl)pyridin-2-yl]piperazine, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
8P11
 
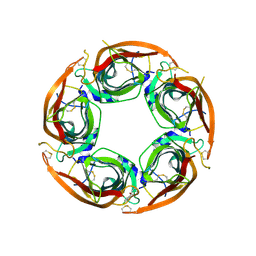 | | X-ray structure of acetylcholine-binding protein (AChBP) in complex with FL003044. | | Descriptor: | 4-(4-chlorophenyl)piperidin-4-ol, Acetylcholine-binding protein, CHLORIDE ION, ... | | Authors: | Cederfelt, D, Boronat, P, Dobritzsch, D, Hennig, S, Fitzgerald, E.A, de Esch, I.J.P, Danielson, U.H. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Elucidating the regulation of ligand gated ion channels via biophysical studies of ligand-induced conformational dynamics of acetylcholine binding proteins
To Be Published
|
|
8P10
 
 | |
8P0Y
 
 | |
8P0T
 
 | |
8P0Q
 
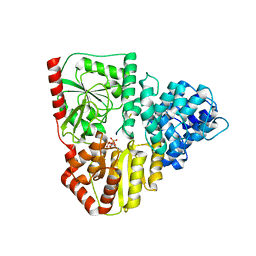 | | Crystal structure of AaNGT complexed to UDP and a peptide | | Descriptor: | Adhesin, PHE-GLY-ASN-TRP-THR-THR, URIDINE-5'-DIPHOSPHATE | | Authors: | Piniello, B, Macias-Leon, J, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for bacterial N-glycosylation by a soluble HMW1C-like N-glycosyltransferase.
Nat Commun, 14, 2023
|
|
8P0P
 
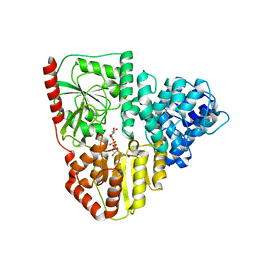 | | Crystal structure of AaNGT complexed to UDP-2F-Glucose | | Descriptor: | Adhesin, URIDINE-5'-DIPHOSPHATE, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE | | Authors: | Piniello, B, Macias-Leon, J, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Molecular basis for bacterial N-glycosylation by a soluble HMW1C-like N-glycosyltransferase.
Nat Commun, 14, 2023
|
|
8P0O
 
 | | Crystal structure of AaNGT complexed to UDP-Gal | | Descriptor: | 1,2-ETHANEDIOL, Adhesin, GALACTOSE-URIDINE-5'-DIPHOSPHATE | | Authors: | Pinello, B, Macias-Leon, J, Rovira, C, Hurtado-Guerrero, R. | | Deposit date: | 2023-05-10 | | Release date: | 2023-09-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular basis for bacterial N-glycosylation by a soluble HMW1C-like N-glycosyltransferase.
Nat Commun, 14, 2023
|
|
8P0M
 
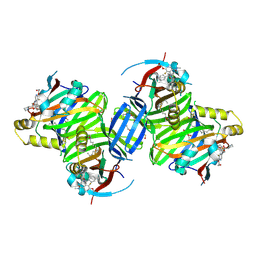 | | Crystal structure of TEAD3 in complex with IAG933 | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 4-[(2~{S})-5-chloranyl-6-fluoranyl-2-phenyl-2-[(2~{S})-pyrrolidin-2-yl]-3~{H}-1-benzofuran-4-yl]-5-fluoranyl-6-(2-hydroxyethyloxy)-~{N}-methyl-pyridine-3-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Scheufler, C, Villard, F, Chau, S. | | Deposit date: | 2023-05-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.961 Å) | | Cite: | Direct and selective pharmacological disruption of the YAP-TEAD interface by IAG933 inhibits Hippo-dependent and RAS-MAPK-altered cancers.
Nat Cancer, 2024
|
|
8P0F
 
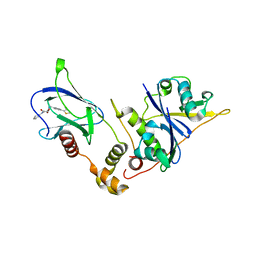 | | Crystal structure of the VCB complex with compound 1. | | Descriptor: | (3~{R},5~{R})-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-5-oxidanyl-2-oxidanylidene-1-pyridin-2-yl-piperidine-3-carboxamide, CHLORIDE ION, Elongin-B, ... | | Authors: | Bader, G, Boettcher, J, Wolkerstorfer, B. | | Deposit date: | 2023-05-10 | | Release date: | 2023-05-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Drugit: Crowd-sourcing molecular design of non-peptidic VHL binders
Chemrxiv, 2023
|
|
8P0D
 
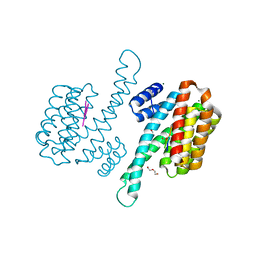 | | Human 14-3-3 sigma in complex with human MDM2 peptide | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Roversi, P, Ward, J, Doveston, R, Kwon, H, Romartinez Alonso, B. | | Deposit date: | 2023-05-10 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Characterizing the protein-protein interaction between MDM2 and 14-3-3 sigma ; proof of concept for small molecule stabilization.
J.Biol.Chem., 300, 2024
|
|
8OZZ
 
 | |
8OZS
 
 | | Populus tremula stable protein 1 with N-terminal binding peptide extension with hemin | | Descriptor: | Stable protein 1 | | Authors: | Sklyar, J, Zeibaq, Y, Bachar, O, Yehezkeli, O, Adir, N. | | Deposit date: | 2023-05-09 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Bioengineered Stable Protein 1-Hemin Complex with Enhanced Peroxidase-Like Catalytic Properties
Small Science, 2023
|
|
8OZQ
 
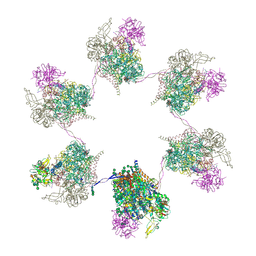 | | In situ subtomogram average of Prototype Foamy Virus Env hexamer of trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Calcraft, T, Nans, A, Rosenthal, P.B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | In situ cryoEM structures of PFV
To Be Published
|
|
8OZP
 
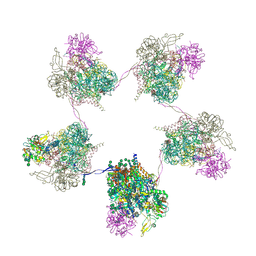 | | In situ subtomogram average of Prototype Foamy Virus Env pentamer of trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Calcraft, T, Nans, A, Rosenthal, P.B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | In situ cryoEM structures of PFV
To Be Published
|
|
8OZN
 
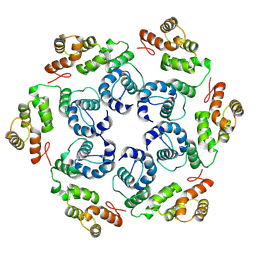 | |
8OZM
 
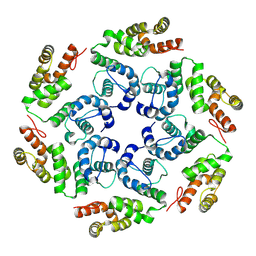 | |
8OZL
 
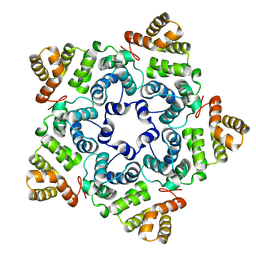 | |
8OZK
 
 | |
