8UH6
 
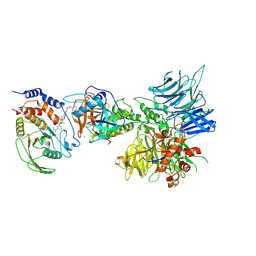 | | Degrader-induced complex between PTPN2 and CRBN-DDB1 | | Descriptor: | (5P)-3-(carboxymethoxy)-4-chloro-5-(3-{[(4S)-1-({3-[(4-{1-[(3R)-2,6-dioxopiperidin-3-yl]-3-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl}piperidine-1-carbonyl)amino]phenyl}methanesulfonyl)-2,2-dimethylpiperidin-4-yl]amino}phenyl)thiophene-2-carboxylic acid, DNA damage-binding protein 1, Protein cereblon, ... | | Authors: | Catalano, C, Bratkowski, M, Scapin, G, Hao, Q. | | Deposit date: | 2023-10-06 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanistic insights into a heterobifunctional degrader-induced PTPN2/N1 complex.
Commun Chem, 7, 2024
|
|
7L5C
 
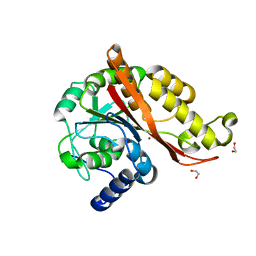 | | Structure of copper bound MEMO1 | | Descriptor: | 1,2-ETHANEDIOL, COPPER (I) ION, GLYCEROL, ... | | Authors: | Boniecki, M.T, Uhlemann, E.E, Dmitriev, O.Y. | | Deposit date: | 2020-12-21 | | Release date: | 2022-01-19 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | MEMO1 binds iron and modulates iron homeostasis in cancer cells.
Elife, 13, 2024
|
|
6V75
 
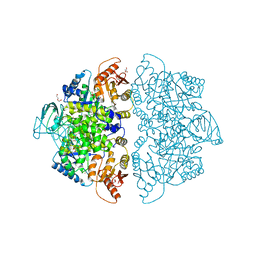 | | Crystal Structure of Human PKM2 in Complex with L-aspartate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Nandi, S, Dey, M. | | Deposit date: | 2019-12-07 | | Release date: | 2020-03-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Biochemical and structural insights into how amino acids regulate pyruvate kinase muscle isoform 2.
J.Biol.Chem., 295, 2020
|
|
6S7C
 
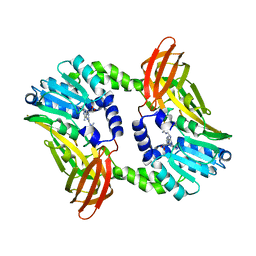 | | Crystal structure of CARM1 in complex with inhibitor UM079 | | Descriptor: | 1-[3-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(3-azanylpropyl)amino]propyl]guanidine, Histone-arginine methyltransferase CARM1 | | Authors: | Gunnell, E.A, Muhsen, U, Dowden, J, Dreveny, I. | | Deposit date: | 2019-07-04 | | Release date: | 2020-03-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and biochemical evaluation of bisubstrate inhibitors of protein arginine N-methyltransferases PRMT1 and CARM1 (PRMT4).
Biochem.J., 477, 2020
|
|
8S76
 
 | | Soluble epoxide hydrolase in complex with PROTAC JSF67 | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional epoxide hydrolase 2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kumar, A, Schoenfeld, J, Hiesinger, K, Lillich, F, Proschak, E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-02-29 | | Release date: | 2025-03-12 | | Last modified: | 2025-09-24 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure-Based Design of PROTACS for the Degradation of Soluble Epoxide Hydrolase.
J.Med.Chem., 68, 2025
|
|
6G2E
 
 | | X-ray structure of NSD3-PWWP1 in complex with compound 13 | | Descriptor: | Histone-lysine N-methyltransferase NSD3, [3,5-dimethyl-4-(1-methyl-5-pyridin-4-yl-imidazol-4-yl)phenyl]methanamine | | Authors: | Boettcher, J, Muellauer, B.J, Weiss-Puxbaum, A, Zoephel, A. | | Deposit date: | 2018-03-23 | | Release date: | 2019-06-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-based discovery of a chemical probe for the PWWP1 domain of NSD3.
Nat.Chem.Biol., 15, 2019
|
|
8ZWS
 
 | | Mtb. MazF-mt3 toxin in compleex with its antitoxin fragmant | | Descriptor: | Antitoxin MazE6, Endoribonuclease MazF6 | | Authors: | Xie, W, Jiang, P. | | Deposit date: | 2024-06-13 | | Release date: | 2025-06-11 | | Last modified: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Long Dynamic beta 1-beta 2 Loops in M. tb MazF Toxins Affect the Interaction Modes and Strengths of the Toxin-Antitoxin Pairs.
Int J Mol Sci, 25, 2024
|
|
1JV6
 
 | | BACTERIORHODOPSIN D85S/F219L DOUBLE MUTANT AT 2.00 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Rouhani, S, Cartailler, J.-P, Facciotti, M.T, Walian, P, Needleman, R, Lanyi, J.K, Glaeser, R.M, Luecke, H. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the D85S mutant of bacteriorhodopsin: model of an O-like photocycle intermediate.
J.Mol.Biol., 313, 2001
|
|
8UP2
 
 | |
8UHK
 
 | |
5I7Y
 
 | | BRD9 in complex with Cpd4 ((E)-3-(6-(but-2-en-1-yl)-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl)-N,N-dimethylbenzamide) | | Descriptor: | 3-{6-[(2E)-but-2-en-1-yl]-7-oxo-6,7-dihydro-1H-pyrrolo[2,3-c]pyridin-4-yl}-N,N-dimethylbenzamide, Bromodomain-containing protein 9 | | Authors: | Murray, J.M. | | Deposit date: | 2016-02-18 | | Release date: | 2016-10-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.4514 Å) | | Cite: | Diving into the Water: Inducible Binding Conformations for BRD4, TAF1(2), BRD9, and CECR2 Bromodomains.
J.Med.Chem., 59, 2016
|
|
9HL5
 
 | | Crystal structure of halo-tolerant PETase from marine metagenome (HaloPETase1) | | Descriptor: | 1,2-ETHANEDIOL, Alpha/beta hydrolase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Turak, O, Kriegel, M, Hocker, B. | | Deposit date: | 2024-12-04 | | Release date: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | A third type of PETase from the marine Halopseudomonas lineage.
Protein Sci., 34, 2025
|
|
8UOH
 
 | | Crystal structure of human NUAK1-MARK3 kinase domain chimera bound with small molecule inhibitor #10 | | Descriptor: | (6P)-6-[(4S)-imidazo[1,2-a]pyridin-3-yl]-4-[(1R)-1-phenylethyl]-2H-pyrido[3,2-b][1,4]oxazin-3(4H)-one, 1,2-ETHANEDIOL, MAP/microtubule affinity-regulating kinase 3, ... | | Authors: | Delker, S.L, Abendroth, J. | | Deposit date: | 2023-10-19 | | Release date: | 2024-10-23 | | Last modified: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of UCB9386: A Potent, Selective, and Brain-Penetrant Nuak1 Inhibitor Suitable for In Vivo Pharmacological Studies.
J.Med.Chem., 67, 2024
|
|
8UFZ
 
 | | Human PU.1 ETS-Domain (165-270) Bound to d(AATAAAAGCGGAAGTG) in Ternary Complex with DB1976 | | Descriptor: | (2M,2'M)-2,2'-(selenophene-2,5-diyl)di(1H-benzimidazole-6-carboximidamide), DNA (5'-D(*AP*AP*TP*AP*AP*AP*AP*GP*CP*GP*GP*AP*AP*GP*TP*G)-3'), DNA (5'-D(*TP*CP*AP*CP*TP*TP*CP*CP*GP*CP*TP*TP*TP*TP*AP*T)-3'), ... | | Authors: | Terrell, J.R, Poon, G.M.K, Wilson, W.D. | | Deposit date: | 2023-10-05 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.06 Å) | | Cite: | Structural studies on the PU.1 inhibitory mechanism by diamidine minor groove binders
To Be Published
|
|
3W92
 
 | | Crystal Structure Analysis of the synthetic GCN4 Thioester coiled coil peptide | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, PARA ACETAMIDO BENZOIC ACID, ... | | Authors: | Shahar, A, Zarivach, R, Ashkenasy, G. | | Deposit date: | 2013-03-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A high-resolution structure that provides insight into coiled-coil thiodepsipeptide dynamic chemistry
Angew.Chem.Int.Ed.Engl., 52, 2013
|
|
7LEO
 
 | | C1B domain of Protein kinase C in complex with diacylglycerol-lactone (AJH-836) and 1,2-diheptanoyl-sn-glycero-3-phosphocholine | | Descriptor: | (4S,7R)-7-(heptanoyloxy)-4-hydroxy-N,N,N-trimethyl-10-oxo-3,5,9-trioxa-4-phosphahexadecan-1-aminium 4-oxide, Protein kinase C delta type, ZINC ION, ... | | Authors: | Katti, S.S, Krieger, I. | | Deposit date: | 2021-01-14 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural anatomy of Protein Kinase C C1 domain interactions with diacylglycerol and other agonists.
Nat Commun, 13, 2022
|
|
5WQ3
 
 | |
6GBL
 
 | | Repertoires of functionally diverse enzymes through computational design at epistatic active-site positions | | Descriptor: | 1,2-ETHANEDIOL, CACODYLATE ION, FORMIC ACID, ... | | Authors: | Khersonsky, O, Lipsh, R, Avizemer, Z, Goldsmith, M, Ashani, Y, Leader, H, Dym, O, Rogotner, S, Trudeau, D, Tawfik, D.S, Fleishman, S.J. | | Deposit date: | 2018-04-15 | | Release date: | 2018-10-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Automated Design of Efficient and Functionally Diverse Enzyme Repertoires.
Mol. Cell, 72, 2018
|
|
7Q9N
 
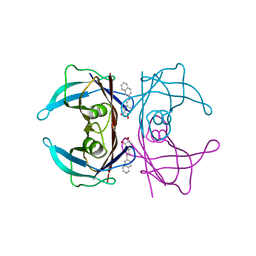 | | Transthyretin complexed with (E)-4-(2-(naphthalen-2-yl)vinyl)benzene-1,2-diol | | Descriptor: | 4-[(~{E})-2-naphthalen-2-ylethenyl]benzene-1,2-diol, Transthyretin | | Authors: | Derbyshire, D.J, Hammarstrom, P, von Castelmur, E, Begum, A. | | Deposit date: | 2021-11-12 | | Release date: | 2022-11-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Transthyretin Binding Mode Dichotomy of Fluorescent trans -Stilbene Ligands.
Acs Chem Neurosci, 14, 2023
|
|
1JV7
 
 | | BACTERIORHODOPSIN O-LIKE INTERMEDIATE STATE OF THE D85S MUTANT AT 2.25 ANGSTROM RESOLUTION | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, Bacteriorhodopsin, RETINAL | | Authors: | Rouhani, S, Cartailler, J.-P, Facciotti, M.T, Walian, P, Needleman, R, Lanyi, J.K, Glaeser, R.M, Luecke, H. | | Deposit date: | 2001-08-28 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the D85S mutant of bacteriorhodopsin: model of an O-like photocycle intermediate.
J.Mol.Biol., 313, 2001
|
|
8EJB
 
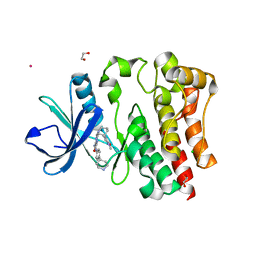 | |
6PRG
 
 | | SBP RafE in complex with stachyose | | Descriptor: | 1,2-ETHANEDIOL, ABC transporter sugar-binding protein, CITRIC ACID, ... | | Authors: | Meier, E.P.W, Boraston, A.B. | | Deposit date: | 2019-07-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular analysis of an enigmaticStreptococcus pneumoniaevirulence factor: The raffinose-family oligosaccharide utilization system.
J.Biol.Chem., 294, 2019
|
|
6VFQ
 
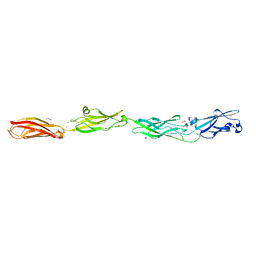 | | Crystal structure of monomeric human protocadherin 10 EC1-EC4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
6VFV
 
 | | Crystal structure of human protocadherin 8 EC5-EC6 | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
3KFL
 
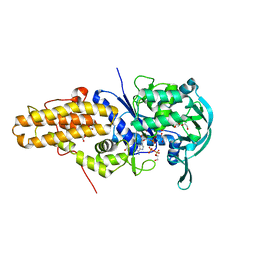 | | Leishmania major methionyl-tRNA synthetase in complex with methionyladenylate and pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Larson, E.T, Merritt, E.A, Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2009-10-27 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Leishmania major methionyl-tRNA synthetase in complex with intermediate products methionyladenylate and pyrophosphate.
Biochimie, 93, 2011
|
|
