1REZ
 
 | | HUMAN LYSOZYME-N-ACETYLLACTOSAMINE COMPLEX | | Descriptor: | GLYCEROL, LYSOZYME, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Origin of carbohydrate recognition specificity of human lysozyme revealed by affinity labeling.
Biochemistry, 35, 1996
|
|
3ORY
 
 | | Crystal structure of Flap endonuclease 1 from hyperthermophilic archaeon Desulfurococcus amylolyticus | | Descriptor: | PHOSPHATE ION, flap endonuclease 1 | | Authors: | Mase, T, Kubota, K, Miyazono, K, Kawarabayashii, Y, Tanokura, M. | | Deposit date: | 2010-09-08 | | Release date: | 2011-02-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of flap endonuclease 1 from the hyperthermophilic archaeon Desulfurococcus amylolyticus
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4YAL
 
 | | Reduced CYPOR with 2'-AMP | | Descriptor: | ADENOSINE-2'-MONOPHOSPHATE, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
1GK1
 
 | | Structure-based prediction of modifications in glutarylamidase to allow single-step enzymatic production of 7-aminocephalosporanic acid from cephalosporin C | | Descriptor: | CEPHALOSPORIN ACYLASE, GLYCEROL | | Authors: | Fritz-Wolf, K, Koller, K.P, Lange, G, Liesum, A, Sauber, K, Schreuder, H, Aretz, W, Kabsch, W. | | Deposit date: | 2001-08-07 | | Release date: | 2002-01-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-Based Prediction of Modifications in Glutarylamidase to Allow Single-Step Enzymatic Production of 7-Aminocephalosporanic Acid from Cephalosporin C.
Protein Sci., 11, 2002
|
|
3P6N
 
 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8-Dans | | Descriptor: | ADAMANTANE-1-CARBOXYLIC ACID-5-DIMETHYLAMINO-NAPHTHALENE-1-SULFONYLAMINO-OCTYL-AMIDE, Camphor 5-monooxygenase, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-10-11 | | Release date: | 2010-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8-Dans
To be Published
|
|
1R3N
 
 | | Crystal structure of beta-alanine synthase from Saccharomyces kluyveri | | Descriptor: | BETA-AMINO ISOBUTYRATE, ZINC ION, beta-alanine synthase | | Authors: | Lundgren, S, Gojkovic, Z, Piskur, J, Dobritzsch, D. | | Deposit date: | 2003-10-02 | | Release date: | 2003-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Yeast beta-Alanine Synthase Shares a Structural Scaffold and Origin with Dizinc-dependent Exopeptidases
J.Biol.Chem., 278, 2003
|
|
4Y9R
 
 | | rat CYPOR mutant - G141del | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Kim, J.J.P. | | Deposit date: | 2015-02-17 | | Release date: | 2016-03-02 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mutants of Cytochrome P450 Reductase Lacking Either Gly-141 or Gly-143 Destabilize Its FMN Semiquinone.
J.Biol.Chem., 291, 2016
|
|
5IDQ
 
 | |
4YA0
 
 | |
4M80
 
 | | The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans at 1.85A resolution | | Descriptor: | EXO-1,3-BETA-GLUCANASE | | Authors: | Nakatani, Y, Cutfield, S.M, Larsen, D.S, Cutfield, J.F. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Major Change in Regiospecificity for the Exo-1,3-beta-glucanase from Candida albicans following Its Conversion to a Glycosynthase.
Biochemistry, 53, 2014
|
|
3OVM
 
 | | X-ray Structural study of quinone reductase II inhibition by compounds with micromolar to nanomolar range IC50 values | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ribosyldihydronicotinamide dehydrogenase [quinone], ZINC ION, ... | | Authors: | Pegan, S.D, Sturdy, M, Ferry, G, Delagrange, P, Boutin, J.A. | | Deposit date: | 2010-09-16 | | Release date: | 2011-05-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | X-ray structural studies of quinone reductase 2 nanomolar range inhibitors.
Protein Sci., 20, 2011
|
|
4YC4
 
 | | Crystal structure of phosphatidyl inositol 4-kinase II alpha in complex with nucleotide analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase type 2-alpha,Lysozyme,Phosphatidylinositol 4-kinase type 2-alpha, [(1S,3S,4S)-3-(6-amino-9H-purin-9-yl)bicyclo[2.2.1]hept-1-yl]methanol | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2015-02-19 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The high-resolution crystal structure of phosphatidylinositol 4-kinase II beta and the crystal structure of phosphatidylinositol 4-kinase II alpha containing a nucleoside analogue provide a structural basis for isoform-specific inhibitor design.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1GL9
 
 | |
4XZ7
 
 | | Crystal structure of a TGase | | Descriptor: | Putative uncharacterized protein | | Authors: | Yu, J, Ge, J, Yang, M. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-24 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Functional and Structural Characterization of the Antiphagocytic Properties of a Novel Transglutaminase from Streptococcus suis
J.Biol.Chem., 290, 2015
|
|
4MAE
 
 | | Methanol dehydrogenase from Methylacidiphilum fumariolicum SolV | | Descriptor: | CERIUM (III) ION, Methanol dehydrogenase, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Pol, A.J, Barends, T.R.M, Dietl, A, Khadem, A.F, Eygenstein, J, Jetten, M.S.M, Op den Camp, H.J.M. | | Deposit date: | 2013-08-16 | | Release date: | 2013-09-18 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rare earth metals are essential for methanotrophic life in volcanic mudpots.
ENVIRON.MICROBIOL., 16, 2014
|
|
3OY3
 
 | | Crystal structure of ABL T315I mutant kinase domain bound with a DFG-out inhibitor AP24589 | | Descriptor: | 5-[(5-{[4-{[4-(2-hydroxyethyl)piperazin-1-yl]methyl}-3-(trifluoromethyl)phenyl]carbamoyl}-2-methylphenyl)ethynyl]-1-methyl-1H-imidazole-2-carboxamide, Tyrosine-protein kinase ABL1 | | Authors: | Zhou, T, Commodore, L, Huang, W.S, Wang, Y, Thomas, M, Keats, J, Xu, Q, Rivera, V, Shakespeare, W.C, Clackson, T, Dalgarno, D.C, Zhu, X. | | Deposit date: | 2010-09-22 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural Mechanism of the Pan-BCR-ABL Inhibitor Ponatinib (AP24534): Lessons for Overcoming Kinase Inhibitor Resistance.
Chem.Biol.Drug Des., 77, 2011
|
|
1GET
 
 | | ANATOMY OF AN ENGINEERED NAD-BINDING SITE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mittl, P.R.E, Schulz, G.E. | | Deposit date: | 1994-01-18 | | Release date: | 1994-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Anatomy of an engineered NAD-binding site.
Protein Sci., 3, 1994
|
|
1GHS
 
 | |
1RRS
 
 | | MutY adenine glycosylase in complex with DNA containing an abasic site | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*T)-3', CALCIUM ION, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2003-12-08 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
1GER
 
 | |
1GLH
 
 | | CATION BINDING TO A BACILLUS (1,3-1,4)-BETA-GLUCANASE. GEOMETRY, AFFINITY AND EFFECT ON PROTEIN STABILITY | | Descriptor: | 1,3-1,4-BETA-GLUCANASE, SODIUM ION | | Authors: | Keitel, T, Heinemann, U. | | Deposit date: | 1994-11-25 | | Release date: | 1995-02-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cation binding to a Bacillus (1,3-1,4)-beta-glucanase. Geometry, affinity and effect on protein stability
Eur.J.Biochem., 222, 1994
|
|
1GBG
 
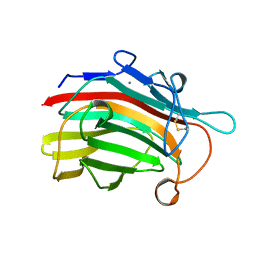 | | BACILLUS LICHENIFORMIS BETA-GLUCANASE | | Descriptor: | (1,3-1,4)-BETA-D-GLUCAN 4 GLUCANOHYDROLASE, CALCIUM ION | | Authors: | Hahn, M, Heinemann, U. | | Deposit date: | 1995-08-25 | | Release date: | 1995-12-07 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Bacillus licheniformis 1,3-1,4-beta-D-glucan 4-glucanohydrolase at 1.8 A resolution.
FEBS Lett., 374, 1995
|
|
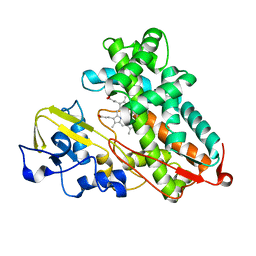
3P6R
 
 | |
1FWJ
 
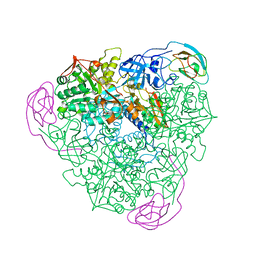 | | KLEBSIELLA AEROGENES UREASE, NATIVE | | Descriptor: | NICKEL (II) ION, UREASE | | Authors: | Pearson, M.A, Karplus, P.A. | | Deposit date: | 1997-04-23 | | Release date: | 1997-10-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of Cys319 variants and acetohydroxamate-inhibited Klebsiella aerogenes urease.
Biochemistry, 36, 1997
|
|
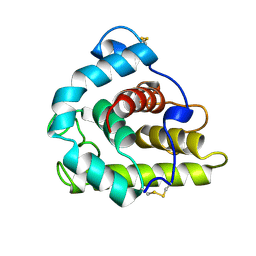
1GBS
 
 | |
