1QK3
 
 | | TOXOPLASMA GONDII HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE GMP COMPLEX | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, HYPOXANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Heroux, A, White, E.L, Ross, L.J, Borhani, D.W. | | Deposit date: | 1999-07-09 | | Release date: | 1999-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structures of the Toxoplasma Gondii Hypoxanthine-Guanine Phosphoribosyltransferase Gmp and -Imp Complexes: Comparison of Purine Binding Interactions with the Xmp Complex
Biochemistry, 38, 1999
|
|
1QM6
 
 | | Closed form of Clostridium perfringens alpha-toxin strain NCTC8237 | | Descriptor: | PHOSPHOLIPASE C, ZINC ION | | Authors: | Naylor, C.E, Miller, J, Titball, R.W, Basak, A.K. | | Deposit date: | 1999-09-21 | | Release date: | 1999-09-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Characterisation of the Calcium-Binding C-Terminal Domain of Clostridium Perfringens Alpha-Toxin
J.Mol.Biol., 294, 1999
|
|
1QJP
 
 | |
7EK9
 
 | | Crystal structure of apo streptavidin at cryogenic temperature | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, Streptavidin | | Authors: | DeMirci, H, Ayan, E, Destan, E, Ertem, F.B. | | Deposit date: | 2021-04-04 | | Release date: | 2021-09-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structure of apo streptavidin at cryogenic temperature
To Be Published
|
|
1TAC
 
 | |
1SSD
 
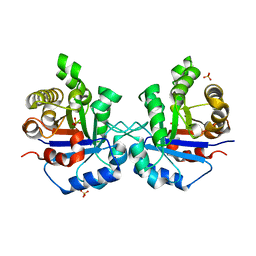 | | Understanding protein lids: Structural analysis of active hinge mutants in triosephosphate isomerase | | Descriptor: | SULFATE ION, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-24 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
7EFX
 
 | | Crystal Structure of human PIN1 complexed with covalent inhibitor | | Descriptor: | 4-((5-bromofuran-2-yl)methyl)-8-(2-chloroacetyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J, Zhu, R, Pei, Y. | | Deposit date: | 2021-03-23 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
1ST7
 
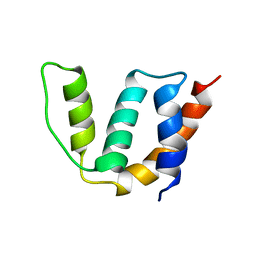 | | Solution structure of Acyl Coenzyme A Binding Protein from yeast | | Descriptor: | Acyl-CoA-binding protein | | Authors: | Teilum, K, Thormann, T, Caterer, N.R, Poulsen, H.I, Jensen, P.H, Knudsen, J, Kragelund, B.B, Poulsen, F.M. | | Deposit date: | 2004-03-25 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Different secondary structure elements as scaffolds for protein folding transition states of two homologous four-helix bundles
Proteins, 59, 2005
|
|
7E3D
 
 | | Crystal structure of human acetylcholinesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Acetylcholinesterase | | Authors: | Dileep, K.V, Ihara, K, Mishima-Tsumagari, C, Kukimoto-Niino, M, Yonemochi, M, Hanada, K, Shirouzu, M, Zhang, K.Y.J. | | Deposit date: | 2021-02-08 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Int.J.Biol.Macromol., 210, 2022
|
|
7E3H
 
 | | Crystal structure of human acetylcholinesterase in complex with donepezil | | Descriptor: | 1-BENZYL-4-[(5,6-DIMETHOXY-1-INDANON-2-YL)METHYL]PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholinesterase | | Authors: | Dileep, K.V, Ihara, K, Mishima-Tsumagari, C, Kukimoto-Niino, M, Yonemochi, M, Hanada, K, Shirouzu, M, Zhang, K.Y.J. | | Deposit date: | 2021-02-08 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Int.J.Biol.Macromol., 210, 2022
|
|
7EKV
 
 | | Crystal Structure of human Pin1 complexed with a covalent inhibitor | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-((5-phenylfuran-2-yl)methyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-04-06 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
7EFJ
 
 | | Crystal Structure Analysis of human PIN1 | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, 8-(2-chloroacetyl)-4-(furan-2-ylmethyl)-1-thia-4,8-diazaspiro[4.5]decan-3-one, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 | | Authors: | Liu, L, Li, J. | | Deposit date: | 2021-03-21 | | Release date: | 2022-02-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Computational and Structure-Based Development of High Potent Cell-Active Covalent Inhibitor Targeting the Peptidyl-Prolyl Isomerase NIMA-Interacting-1 (Pin1).
J.Med.Chem., 65, 2022
|
|
1TER
 
 | |
1SW0
 
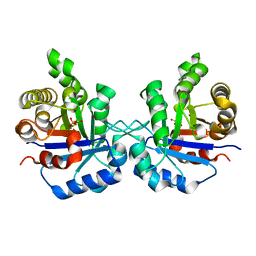 | | Triosephosphate isomerase from Gallus gallus, loop 6 hinge mutant K174L, T175W | | Descriptor: | 2-PHOSPHOGLYCOLIC ACID, Triosephosphate isomerase | | Authors: | Kursula, I, Salin, M, Sun, J, Norledge, B.V, Haapalainen, A.M, Sampson, N.S, Wierenga, R.K. | | Deposit date: | 2004-03-30 | | Release date: | 2004-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Understanding protein lids: structural analysis of active hinge mutants in triosephosphate isomerase
Protein Eng.Des.Sel., 17, 2004
|
|
1THK
 
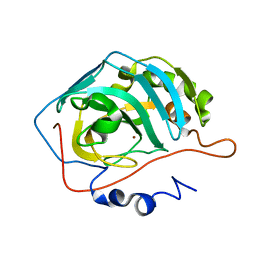 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-06-01 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
1TKT
 
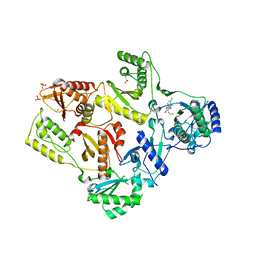 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GW426318 | | Descriptor: | 6-CHLORO-4-(CYCLOHEXYLOXY)-3-PROPYLQUINOLIN-2(1H)-ONE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Hopkins, A.L, Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2004-06-09 | | Release date: | 2004-12-07 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Design of non-nucleoside inhibitors of HIV-1 reverse transcriptase with improved drug resistance properties. 1.
J.Med.Chem., 47, 2004
|
|
1TSX
 
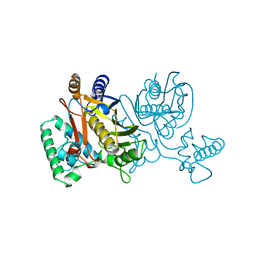 | | THYMIDYLATE SYNTHASE R179E MUTANT | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Finer-Moore, J, Stroud, R.M. | | Deposit date: | 1995-12-05 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Contribution of a salt bridge to binding affinity and dUMP orientation to catalytic rate: mutation of a substrate-binding arginine in thymidylate synthase.
Protein Eng., 9, 1996
|
|
1U3F
 
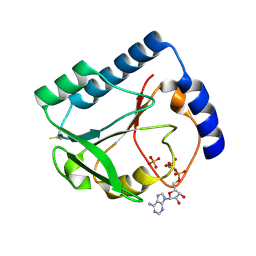 | | Structural and Functional Characterization of a 5,10-Methenyltetrahydrofolate Synthetase from Mycoplasma pneumoniae (GI: 13508087) | | Descriptor: | 5,10-Methenyltetrahydrofolate Synthetase, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Chen, S, Yakunin, A.F, Proudfoot, M, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-07-21 | | Release date: | 2004-12-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of methenyltetrahydrofolate synthetase from Mycoplasma pneumoniae (GI: 13508087) at 2.2 A resolution
Proteins, 56, 2004
|
|
7DVM
 
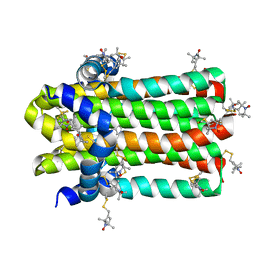 | | DgkA structure in E.coli lipid bilayer | | Descriptor: | Diacylglycerol kinase | | Authors: | Li, J, Yang, J. | | Deposit date: | 2021-01-13 | | Release date: | 2022-04-13 | | Last modified: | 2023-09-27 | | Method: | SOLID-STATE NMR | | Cite: | Structure of membrane diacylglycerol kinase in lipid bilayers.
Commun Biol, 4, 2021
|
|
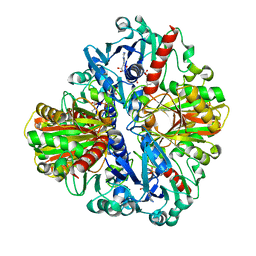
1U8F
 
 | |
1U7P
 
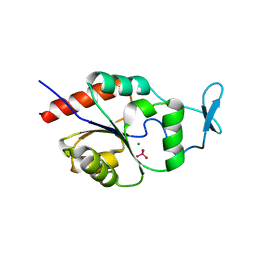 | | X-ray Crystal Structure of the Hypothetical Phosphotyrosine Phosphatase MDP-1 of the Haloacid Dehalogenase Superfamily | | Descriptor: | MAGNESIUM ION, TUNGSTATE(VI)ION, magnesium-dependent phosphatase-1 | | Authors: | Peisach, E, Selengut, J.D, Dunaway-Mariano, D, Allen, K.N. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray Crystal Structure of the Hypothetical Phosphotyrosine
Phosphatase MDP-1 of the Haloacid Dehalogenase Superfamily
Biochemistry, 43, 2004
|
|
1U75
 
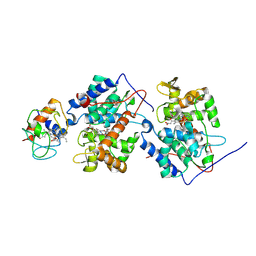 | |
7DYK
 
 | |
7DYI
 
 | |
7DYJ
 
 | |
