4QWJ
 
 | | yCP beta5-A49T-mutant in complex with carfilzomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4R5Y
 
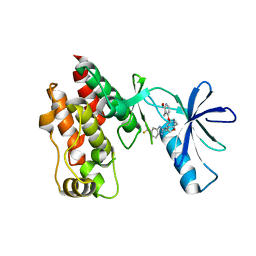 | | The complex structure of Braf V600E kinase domain with a novel Braf inhibitor | | Descriptor: | 5-({(1R,1aS,6bR)-1-[5-(trifluoromethyl)-1H-benzimidazol-2-yl]-1a,6b-dihydro-1H-cyclopropa[b][1]benzofuran-5-yl}oxy)-3,4-dihydro-1,8-naphthyridin-2(1H)-one, Serine/threonine-protein kinase B-raf | | Authors: | Feng, Y, Peng, H, Zhang, Y, Liu, Y, Wei, M. | | Deposit date: | 2014-08-22 | | Release date: | 2016-02-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | BGB-283, a Novel RAF Kinase and EGFR Inhibitor, Displays Potent Antitumor Activity in BRAF-Mutated Colorectal Cancers.
Mol.Cancer Ther., 14, 2015
|
|
4QZ2
 
 | | yCP beta5-M45I mutant in complex with the epoxyketone inhibitor ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-27 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
7Z7D
 
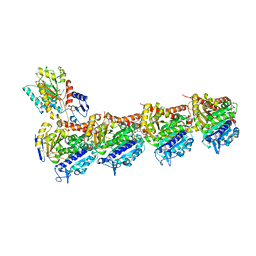 | | Tubulin-Todalam-Vinblastine-complex | | Descriptor: | (2ALPHA,2'BETA,3BETA,4ALPHA,5BETA)-VINCALEUKOBLASTINE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Muehlethaler, T, Milanos, L, Ortega, J.A, Blum, T.B, Gioia, D, Roy, B, Prota, A.E, Cavalli, A, Steinmetz, M.O. | | Deposit date: | 2022-03-15 | | Release date: | 2022-04-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rational Design of a Novel Tubulin Inhibitor with a Unique Mechanism of Action.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
3VQ4
 
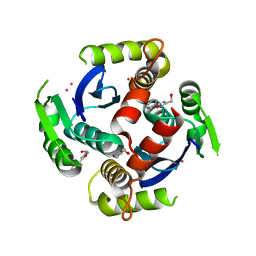 | | Fragments bound to HIV-1 integrase | | Descriptor: | (5-phenyl-1,2-oxazol-3-yl)methanol, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
4RG6
 
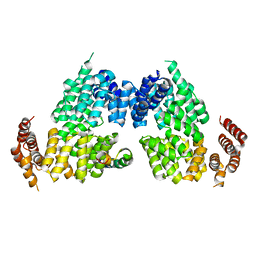 | | Crystal structure of APC3-APC16 complex | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
3VQ6
 
 | | HIV-1 IN core domain in complex with (1-methyl-5-phenyl-1H-pyrazol-4-yl)methanol | | Descriptor: | (1-methyl-5-phenyl-1H-pyrazol-4-yl)methanol, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
4QV0
 
 | | yCP beta5-A49T-A50V-double mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QVY
 
 | | yCP beta5-A49T-mutant in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QWR
 
 | | yCP beta5-C52F mutant in complex with carfilzomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-17 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4R00
 
 | | yCP beta5-C52F mutant in complex with Omuralide | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Omuralide, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-29 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
5Y03
 
 | | Galectin-13/Placental Protein 13 variant R53H crystal structure | | Descriptor: | Galactoside-binding soluble lectin 13 | | Authors: | Wang, Y, Su, J. | | Deposit date: | 2017-07-14 | | Release date: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Galectin-13, a different prototype galectin, does not bind beta-galacto-sides and forms dimers via intermolecular disulfide bridges between Cys-136 and Cys-138.
Sci Rep, 8, 2018
|
|
6MG8
 
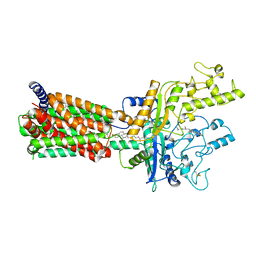 | | Structural basis for cholesterol transport-like activity of the Hedgehog receptor Patched | | Descriptor: | CHOLESTEROL, Protein patched homolog 1 | | Authors: | Zhang, Y, Bulkley, D, Xin, Y, Roberts, K.J, Asarnow, D.E, Sharma, A, Myers, B.R, Cho, W, Cheng, Y, Beachy, P.A. | | Deposit date: | 2018-09-13 | | Release date: | 2018-11-28 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Basis for Cholesterol Transport-like Activity of the Hedgehog Receptor Patched.
Cell, 175, 2018
|
|
5CCC
 
 | |
4QVN
 
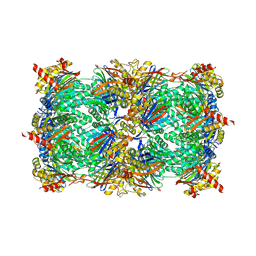 | | yCP beta5-M45V mutant in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4QWF
 
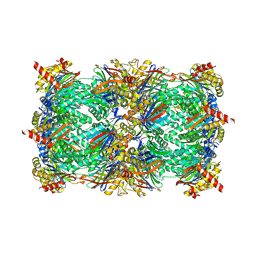 | | yCP beta5-M45I mutant in complex with carfilzomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
5XZI
 
 | |
4Y3U
 
 | | The structure of phospholamban bound to the calcium pump SERCA1a | | Descriptor: | Cardiac phospholamban, POTASSIUM ION, Sarcoplasmic/endoplasmic reticulum calcium ATPase 1 | | Authors: | Hurley, T.D. | | Deposit date: | 2015-02-10 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | The structural basis for phospholamban inhibition of the calcium pump in sarcoplasmic reticulum.
J. Biol. Chem., 288, 2013
|
|
4RDP
 
 | | Crystal structure of Cmr4 | | Descriptor: | CRISPR system Cmr subunit Cmr4 | | Authors: | Shao, Y, Tang, L, Li, H. | | Deposit date: | 2014-09-19 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Essential Structural and Functional Roles of the Cmr4 Subunit in RNA Cleavage by the Cmr CRISPR-Cas Complex.
Cell Rep, 9, 2014
|
|
3VQC
 
 | | HIV-1 IN core domain in complex with (5-METHYL-3-PHENYL-1,2-OXAZOL-4-YL)METHANOL | | Descriptor: | (5-methyl-3-phenyl-1,2-oxazol-4-yl)methanol, CADMIUM ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
4R3P
 
 | | Crystal structures of EGFR in complex with Mig6 | | Descriptor: | Epidermal growth factor receptor, peptide from ERBB receptor feedback inhibitor 1 | | Authors: | Park, E, Kim, N, Yi, Z, Cho, A, Kim, K, Ficarro, S.B, Park, A, Park, W.Y, Murray, B, Meyerson, M, Beroukim, R, Marto, J.A, Cho, J, Eck, M.J. | | Deposit date: | 2014-08-17 | | Release date: | 2015-08-12 | | Last modified: | 2015-09-16 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Structure and mechanism of activity-based inhibition of the EGF receptor by Mig6.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4RG9
 
 | | Crystal structure of APC3-APC16 complex (Selenomethionine Derivative) | | Descriptor: | Anaphase-promoting complex subunit 16, Cell division cycle protein 27 homolog | | Authors: | Yamaguchi, M, Yu, S, Miller, D.J, Schulman, B.A. | | Deposit date: | 2014-09-29 | | Release date: | 2014-12-24 | | Last modified: | 2017-08-02 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of an APC3-APC16 Complex: Insights into Assembly of the Anaphase-Promoting Complex/Cyclosome.
J.Mol.Biol., 427, 2015
|
|
4Y7P
 
 | | Structure of alkaline D-peptidase from Bacillus cereus | | Descriptor: | Alkaline D-peptidase, THIOCYANATE ION | | Authors: | Nakano, S, Okazaki, S, Ishitsubo, E, Kawahara, N, Komeda, H, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-02-15 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational analysis of peptide recognition mechanism of class-C type penicillin binding protein, alkaline D-peptidase from Bacillus cereus DF4-B
Sci Rep, 5, 2015
|
|
8F2F
 
 | |
3VQE
 
 | | HIV-1 IN core domain in complex with [1-(4-fluorophenyl)-5-methyl-1H-pyrazol-4-yl]methanol | | Descriptor: | CADMIUM ION, CHLORIDE ION, POL polyprotein, ... | | Authors: | Wielens, J, Chalmers, D.K, Parker, M.W, Scanlon, M.J. | | Deposit date: | 2012-03-21 | | Release date: | 2013-01-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Parallel screening of low molecular weight fragment libraries: do differences in methodology affect hit identification?
J Biomol Screen, 18, 2013
|
|
