5GXW
 
 | | Importin and NuMA complex | | Descriptor: | Importin subunit alpha-1, Peptide from Nuclear mitotic apparatus protein 1 | | Authors: | Chang, C.-C, Huang, T.-L, Hsia, K.-C. | | Deposit date: | 2016-09-20 | | Release date: | 2017-10-04 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.394 Å) | | Cite: | Regulation of mitotic spindle assembly factor NuMA by Importin-beta
J. Cell Biol., 216, 2017
|
|
5GJC
 
 | | Zika virus NS3 helicase in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MANGANESE (II) ION, NS3 helicase | | Authors: | Tian, H.L, Ji, X.Y, Yang, X.Y, Zhang, Z.X, Lu, Z.K, Yang, K.L, Chen, C, Zhao, Q, Chi, H, Mu, Z.Y, Xie, W, Wang, Z.F, Lou, H.Q, Yang, H.T, Rao, Z.H. | | Deposit date: | 2016-06-28 | | Release date: | 2016-07-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Structural basis of Zika virus helicase in recognizing its substrates
Protein Cell, 7, 2016
|
|
3KM3
 
 | |
3L0K
 
 | |
5GY8
 
 | |
3KM8
 
 | |
5GJG
 
 | | Crystal structure of human TAK1/TAB1 fusion protein in complex with ligand 4 | | Descriptor: | N-(2-isopropoxy-4-(4-methylpiperazine-1-carbonyl)phenyl)-2-(3-(phenylcarbamoyl)phenyl)thiazole-4-carboxamide, TAK1 kinase - TAB1 chimera fusion protein | | Authors: | Irie, M, Nakamura, M, Fukami, T.A, Matsuura, T, Morishima, K. | | Deposit date: | 2016-06-29 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Development of a Method for Converting a TAK1 Type I Inhibitor into a Type II or c-Helix-Out Inhibitor by Structure-Based Drug Design (SBDD)
Chem.Pharm.Bull., 64, 2016
|
|
5GYC
 
 | |
3KMQ
 
 | | G62S mutant of foot-and-mouth disease virus RNA-polymerase in complex with a template- primer RNA, tetragonal structure | | Descriptor: | 3D polymerase, RNA (5'-R(*GP*GP*CP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*CP*C)-3') | | Authors: | Ferrer-Orta, C, Verdaguer, N, Perez-Luque, R. | | Deposit date: | 2009-11-11 | | Release date: | 2010-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structure of foot-and-mouth disease virus mutant polymerases with reduced sensitivity to ribavirin
J.Virol., 84, 2010
|
|
5GJM
 
 | |
5GYG
 
 | |
5GK0
 
 | |
3L1Z
 
 | |
5GZ2
 
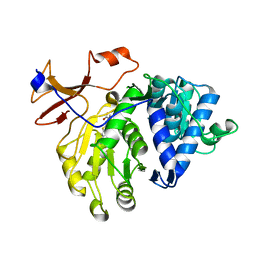 | | luciferase complex with 7-cy-L | | Descriptor: | (4S)-2-[6-(azepan-1-yl)-1,3-benzothiazol-2-yl]-4,5-dihydro-1,3-thiazole-4-carboxylic acid, Luciferin 4-monooxygenase | | Authors: | Su, J, Wang, F, Gu, L. | | Deposit date: | 2016-09-26 | | Release date: | 2017-09-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | luciferase complex with 7-cy-L
To Be Published
|
|
3KNW
 
 | | Crystal structure of a putative transcriptional regulator (TetR/AcrR family member) from putative transcriptional regulator (TetR/AcrR family) | | Descriptor: | 1,2-ETHANEDIOL, Putative transcriptional regulator (TetR/AcrR family) | | Authors: | Nocek, B, Bigelow, L, Cobb, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-11-12 | | Release date: | 2010-01-05 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure of a putative transcriptional regulator (TetR/AcrR family member) from putative transcriptional regulator (TetR/AcrR family)
To be Published
|
|
3L2A
 
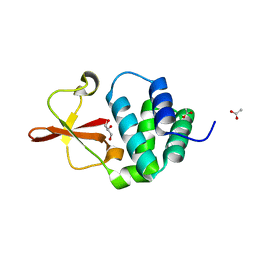 | | Crystal structure of Reston Ebola VP35 interferon inhibitory domain | | Descriptor: | ACETIC ACID, GLYCEROL, Polymerase cofactor VP35 | | Authors: | Leung, D.W, Farahbakhsh, M, Borek, D.M, Prins, K.C, Basler, C.F, Amarasinghe, G.K. | | Deposit date: | 2009-12-14 | | Release date: | 2010-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural and Functional Characterization of Reston Ebola Virus VP35 Interferon Inhibitory Domain.
J.Mol.Biol., 399, 2010
|
|
5GKQ
 
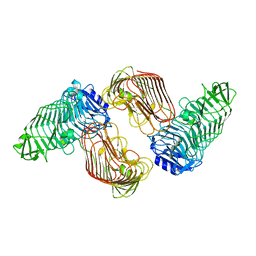 | | Structure of PL6 family alginate lyase AlyGC mutant-R241A | | Descriptor: | AlyGC mutant - R241A, CALCIUM ION, beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid-(1-4)-beta-D-mannopyranuronic acid | | Authors: | Zhang, Y.Z, Wang, P, Xu, F. | | Deposit date: | 2016-07-05 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.565 Å) | | Cite: | Novel Molecular Insights into the Catalytic Mechanism of Marine Bacterial Alginate Lyase AlyGC from Polysaccharide Lyase Family 6
J. Biol. Chem., 292, 2017
|
|
5GZA
 
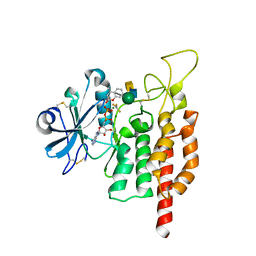 | | protein O-mannose kinase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose, 4-METHYL-2H-CHROMEN-2-ONE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Xiao, J. | | Deposit date: | 2016-09-27 | | Release date: | 2016-12-07 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of protein O-mannose kinase reveals a unique active site architecture
Elife, 5, 2016
|
|
3KOI
 
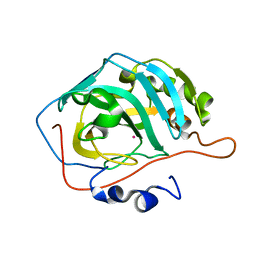 | |
5GL4
 
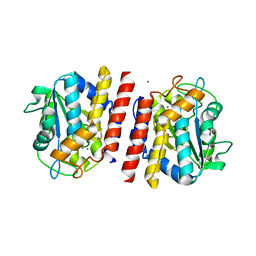 | | Crystal structure of TON_0340 in complex with Mn | | Descriptor: | MANGANESE (II) ION, Uncharacterized protein | | Authors: | Lee, S.G, Sohn, Y.S, Oh, B.H. | | Deposit date: | 2016-07-08 | | Release date: | 2016-12-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of a Highly Conserved Hypothetical Protein TON_0340 as a Probable Manganese-Dependent Phosphatase.
PLoS ONE, 11, 2016
|
|
3L2I
 
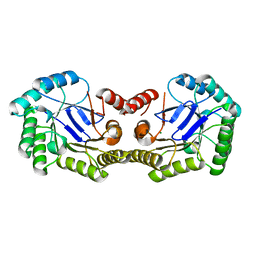 | | 1.85 Angstrom Crystal Structure of the 3-Dehydroquinate Dehydratase (aroD) from Salmonella typhimurium LT2. | | Descriptor: | 3-dehydroquinate dehydratase, MAGNESIUM ION | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-15 | | Release date: | 2009-12-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A conserved surface loop in type I dehydroquinate dehydratases positions an active site arginine and functions in substrate binding.
Biochemistry, 50, 2011
|
|
3KON
 
 | |
5GLJ
 
 | |
5GZU
 
 | | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery | | Descriptor: | Chitinase, PHOSPHATE ION | | Authors: | Itoh, T, Hibi, T, Suzuki, F, Sugimoto, I, Fujiwara, A, Inaka, K, Tanaka, H, Ohta, K, Fujii, Y, Taketo, A, Kimoto, H. | | Deposit date: | 2016-10-01 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Crystal Structure of Chitinase ChiW from Paenibacillus sp. str. FPU-7 Reveals a Novel Type of Bacterial Cell-Surface-Expressed Multi-Modular Enzyme Machinery
PLoS ONE, 11, 2016
|
|
3KOX
 
 | | Crystal Structure of ornithine 4,5 aminomutase in complex with 2,4-diaminobutyrate (Anaerobic) | | Descriptor: | (2S)-2-amino-4-{[(1Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}butanoic acid, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
