6K8N
 
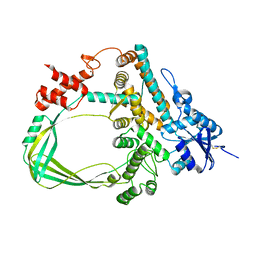 | | Crystal structure of the Sulfolobus solfataricus topoisomerase III | | Descriptor: | ZINC ION, topoisomerase III | | Authors: | Wang, H.Q, Zhang, J.H, Zheng, X, Zheng, Z.F, Dong, Y.H, Huang, L, Gong, Y. | | Deposit date: | 2019-06-13 | | Release date: | 2020-06-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of the Sulfolobus solfataricus topoisomerase III reveal that its C-terminal novel zinc finger part is a unique decatenation domain
To Be Published
|
|
7QQ3
 
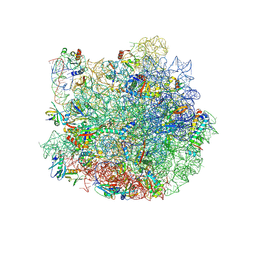 | | Cryo-EM structure of the E.coli 50S ribosomal subunit in complex with the antibiotic Myxovalargin A. | | Descriptor: | 23S ribosomal RNA, 50S ribosomal protein L13, 50S ribosomal protein L14, ... | | Authors: | Koller, T.O, Beckert, B, Wilson, D.N. | | Deposit date: | 2022-01-06 | | Release date: | 2023-01-18 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | The Myxobacterial Antibiotic Myxovalargin: Biosynthesis, Structural Revision, Total Synthesis, and Molecular Characterization of Ribosomal Inhibition.
J.Am.Chem.Soc., 145, 2023
|
|
6XZ7
 
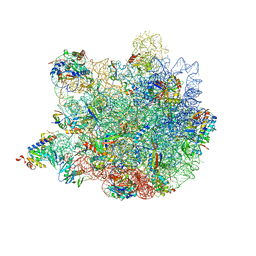 | | E. coli 50S ribosomal subunit in complex with dirithromycin, fMet-Phe-tRNA(Phe) and deacylated tRNA(iMet). | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Pichkur, E.B, Polikanov, Y.S, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2020-02-03 | | Release date: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Insights into the improved macrolide inhibitory activity from the high-resolution cryo-EM structure of dirithromycin bound to theE. coli70S ribosome.
Rna, 26, 2020
|
|
5FP6
 
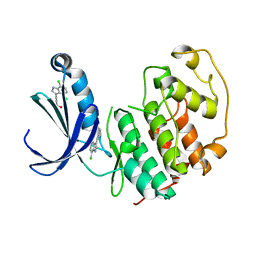 | | Structure of cyclin-dependent kinase 2 with small-molecule ligand 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol (AT17833) in an alternate binding site. | | Descriptor: | 3-(4,7-dichloro-1H-indol-3-yl)prop-2-yn-1-ol, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Jhoti, H, Ludlow, R.F, O'Reilly, M, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
8XXL
 
 | | Cryo-EM structure of the human 40S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8XXM
 
 | | Cryo-EM structure of the human 40S ribosome with PDCD4 and eIF3G | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
8XXN
 
 | | Cryo-EM structure of the human 43S ribosome with PDCD4 | | Descriptor: | 18S rRNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Ye, X, Huang, Z, Li, Y, Wang, M, Cheng, J. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-01 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Human tumor suppressor PDCD4 directly interacts with ribosomes to repress translation.
Cell Res., 34, 2024
|
|
7JSS
 
 | |
8GCJ
 
 | | PCNA | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Vandborg, B.C, Bruning, J.B. | | Deposit date: | 2023-03-01 | | Release date: | 2024-05-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of apo pcna
To Be Published
|
|
7JSW
 
 | |
7EDS
 
 | |
7EEU
 
 | |
7K53
 
 | | Pre-translocation +1-frameshifting(CCC-A) complex (Structure I-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K51
 
 | | Mid-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K50
 
 | | Pre-translocation non-frameshifting(CCA-A) complex (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7Q4K
 
 | | Erythromycin-stalled Escherichia coli 70S ribosome with streptococcal MsrDL nascent chain | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fostier, C.R, Ousalem, F, Soufari, H, Leroy, E.C, Ngo, S, Innis, A, Hashem, Y, Boel, G. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Regulation of the macrolide resistance ABC-F translation factor MsrD.
Nat Commun, 14, 2023
|
|
7JT2
 
 | |
7K00
 
 | | Structure of the Bacterial Ribosome at 2 Angstrom Resolution | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Watson, Z.L, Ward, F.R, Meheust, R, Ad, O, Schepartz, A, Banfield, J.F, Cate, J.H.D. | | Deposit date: | 2020-09-02 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Structure of the bacterial ribosome at 2 angstrom resolution.
Elife, 9, 2020
|
|
8OJ5
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (in-vitro reconstitution) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Peter, J.J, Kulathu, Y, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OHD
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 3 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-21 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
8OJ0
 
 | | 60S ribosomal subunit bound to the E3-UFM1 complex - state 2 (native) | | Descriptor: | 28S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Penchev, I, DaRosa, P.A, Becker, T, Beckmann, R, Kopito, R. | | Deposit date: | 2023-03-23 | | Release date: | 2024-02-21 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | UFM1 E3 ligase promotes recycling of 60S ribosomal subunits from the ER.
Nature, 627, 2024
|
|
7K55
 
 | | Near post-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure III-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
6WYV
 
 | | E. coli 50S ribosome bound to compounds 47 and VS1 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2020-05-13 | | Release date: | 2020-06-17 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
7K54
 
 | | Mid-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure II-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K52
 
 | | Near post-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
