6BHI
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, UNKNOWN ATOM OR ION | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6I9G
 
 | |
7LVW
 
 | | Structure of RSV F in Complex with VHH Cl184 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, F-VHH-Cl184, ... | | Authors: | Hsieh, C.-L, McLellan, J.S. | | Deposit date: | 2021-02-26 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A vulnerable, membrane-proximal site in human respiratory syncytial virus F revealed by a prefusion-specific single-domain antibody.
J.Virol., 95, 2021
|
|
7LVC
 
 | | E. coli DHFR by Native Mn,P,S-SAD at Room Temperature | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Greisman, J.B, Dalton, K.M, Hekstra, D.R. | | Deposit date: | 2021-02-24 | | Release date: | 2021-03-17 | | Last modified: | 2022-08-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Native SAD phasing at room temperature.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6BQP
 
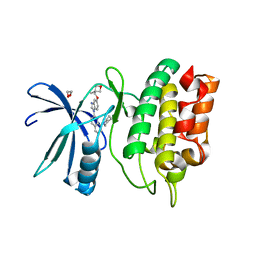 | | Crystal Structure of the Human CAMKK2B in complex with Crenolanib | | Descriptor: | 1,2-ETHANEDIOL, 1-(2-{5-[(3-Methyloxetan-3-yl)methoxy]-1H-benzimidazol-1-yl}quinolin-8-yl)piperidin-4-amine, Calcium/calmodulin-dependent protein kinase kinase 2 | | Authors: | Counago, R.M, de Souza, G.P, dos Reis, C.V, Ramos, P.Z, Drewry, D, Massirer, K.B, Arruda, P, Edwards, A.M, Elkins, J.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-28 | | Release date: | 2017-12-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of the Human CAMKK2B in complex with Crenolanib
To Be Published
|
|
6I5H
 
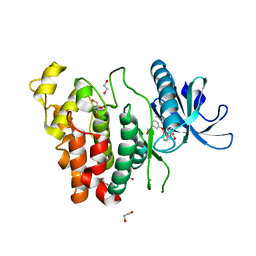 | | Crystal structure of CLK1 in complex with furanopyrimidin VN412 | | Descriptor: | 1,2-ETHANEDIOL, 5-(1-methylpyrazol-4-yl)-3-(3-phenoxyphenyl)furo[3,2-b]pyridine, Dual specificity protein kinase CLK1, ... | | Authors: | Schroeder, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Paruch, K, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Furo[3,2-b]pyridine: A Privileged Scaffold for Highly Selective Kinase Inhibitors and Effective Modulators of the Hedgehog Pathway.
Angew. Chem. Int. Ed. Engl., 58, 2019
|
|
7A0D
 
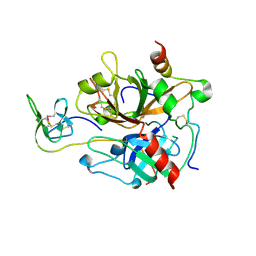 | | The Crystal Structure of Bovine Thrombin in complex with Hirudin (C16U/C28U) at 1.6 Angstroms Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin variant-1, Prothrombin, ... | | Authors: | Hidmi, T, Mousa, R, Pomyalov, S, Lansky, S, Khouri, L, Metanis, N, Shoham, G. | | Deposit date: | 2020-08-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Diselenide crosslinks for enhanced and simplified oxidative protein folding
Commun Chem, 4, 2021
|
|
7LYH
 
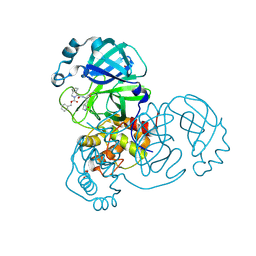 | | Crystal structure of the SARS-CoV-2 (COVID-19) main protease in complex with inhibitor UAWJ9-36-1 | | Descriptor: | 3C-like proteinase, GLYCEROL, benzyl (1S,3aR,6aS)-1-({(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}carbamoyl)hexahydrocyclopenta[c]pyrrole-2(1H)-carboxylate | | Authors: | Sacco, M, Wang, J, Chen, Y. | | Deposit date: | 2021-03-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational Design of Hybrid SARS-CoV-2 Main Protease Inhibitors Guided by the Superimposed Cocrystal Structures with the Peptidomimetic Inhibitors GC-376, Telaprevir, and Boceprevir.
Acs Pharmacol Transl Sci, 4, 2021
|
|
8R0Z
 
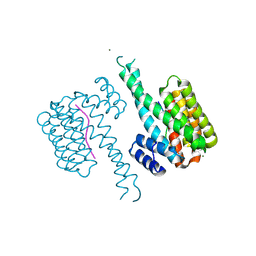 | | 14-3-3 sigma in complex with TAZ peptide and stabilizing fragment TCF199 | | Descriptor: | 14-3-3 protein sigma, MAGNESIUM ION, WW domain-containing transcription regulator protein 1, ... | | Authors: | Centorrino, F, Andlovic, B, Ottmann, C. | | Deposit date: | 2023-11-01 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Fragment-Based Interrogation of the 14-3-3/TAZ Protein-Protein Interaction.
Biochemistry, 63, 2024
|
|
6BK6
 
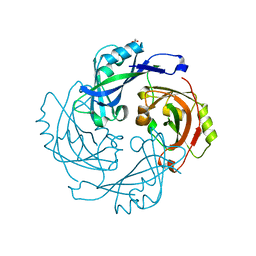 | | Crystal structure of Hendra virus matrix protein | | Descriptor: | ACETATE ION, Hendra virus matrix protein | | Authors: | Liu, Y.C. | | Deposit date: | 2017-11-07 | | Release date: | 2018-04-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Electrostatic Interactions between Hendra Virus Matrix Proteins Are Required for Efficient Virus-Like-Particle Assembly.
J. Virol., 92, 2018
|
|
7LVU
 
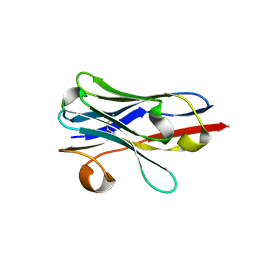 | | Structure of RSV F-directed VHH Cl184 | | Descriptor: | F-VHH-Cl184 | | Authors: | Hsieh, C.-L, McLellan, J.S. | | Deposit date: | 2021-02-26 | | Release date: | 2021-03-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A vulnerable, membrane-proximal site in human respiratory syncytial virus F revealed by a prefusion-specific single-domain antibody.
J.Virol., 95, 2021
|
|
6I5Q
 
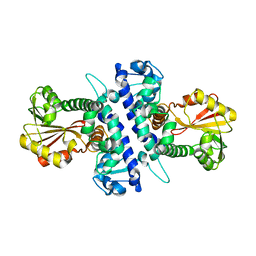 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | O-methyltransferase 1 | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-14 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
7ADV
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ447 (compound 6v) | | Descriptor: | 4-azanyl-~{N}-[[2,4-bis(fluoranyl)phenyl]methyl]-6-[2-(2-morpholin-4-ylethylsulfonyl)ethyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Pye, V.E, Cherepanov, P. | | Deposit date: | 2020-09-16 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | HIV-1 Integrase Inhibitors with Modifications That Affect Their Potencies against Drug Resistant Integrase Mutants.
Acs Infect Dis., 7, 2021
|
|
6I6L
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, Tetrahydrocolumbamine | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
6BHD
 
 | | Crystal structure of SETDB1 with a modified H3 peptide | | Descriptor: | Histone H3.1, Histone-lysine N-methyltransferase SETDB1, SODIUM ION, ... | | Authors: | Qin, S, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-10-30 | | Release date: | 2017-12-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | H3K14ac is linked to methylation of H3K9 by the triple Tudor domain of SETDB1.
Nat Commun, 8, 2017
|
|
6I75
 
 | | Galectin-3C in complex with substituted polyfluoroaryl monothiogalactoside derivative 2 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R},6~{S})-2-(hydroxymethyl)-6-(4-methylphenyl)sulfanyl-4-[4-[2,3,5,6-tetrakis(fluoranyl)-4-oxidanyl-phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3 | | Authors: | Kumar, R, Peterson, K, Nilsson, U.J, Logan, D.T. | | Deposit date: | 2018-11-15 | | Release date: | 2019-01-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.171 Å) | | Cite: | Substituted polyfluoroaryl interactions with an arginine side chain in galectin-3 are governed by steric-, desolvation and electronic conjugation effects.
Org. Biomol. Chem., 17, 2019
|
|
7A6W
 
 | | Structure of the FKBP51FK1 domain in complex with the macrocyclic SAFit analogue 33-(Z) | | Descriptor: | (2S,9S,12R,20Z)-2-cyclohexyl-12-[2-(3,4-dimethoxyphenyl)ethyl]-28,31-dimethoxy-11,18,23,26-tetraoxa-4-azatetracyclo[25.2.2.113,17.04,9]dotriaconta-1(29),13(32),14,16,20,27,30-heptaene-3,10-dione, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Bauder, M, Meyners, C, Purder, P, Merz, S, Voll, A, Heymann, T, Hausch, F. | | Deposit date: | 2020-08-27 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design of High-Affinity Macrocyclic FKBP51 Inhibitors.
J.Med.Chem., 64, 2021
|
|
6BL3
 
 | | Crystal Complex of Cyclooxygenase-2 with indomethacin-butyldiamine-dansyl conjugate | | Descriptor: | 2-[1-(4-chlorobenzene-1-carbonyl)-5-methoxy-2-methyl-1H-indol-3-yl]-N-[4-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)butyl]acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xu, S, Uddin, M.J, Banerjee, S, Marnett, L.J. | | Deposit date: | 2017-11-09 | | Release date: | 2018-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.217 Å) | | Cite: | Fluorescent indomethacin-dansyl conjugates utilize the membrane-binding domain of cyclooxygenase-2 to block the opening to the active site.
J.Biol.Chem., 294, 2019
|
|
7A0F
 
 | | The Crystal Structure of Bovine Thrombin in complex with Hirudin (C22U/C39U) at 2.7 Angstroms Resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hirudin variant-1, Prothrombin, ... | | Authors: | Hidmi, T, Mousa, R, Pomyalov, S, Lansky, S, Khouri, L, Metanis, N, Shoham, G. | | Deposit date: | 2020-08-07 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Diselenide crosslinks for enhanced and simplified oxidative protein folding
Commun Chem, 4, 2021
|
|
7LHX
 
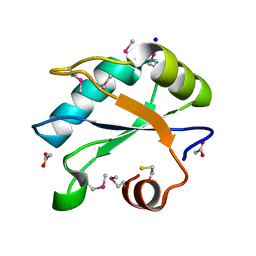 | | Human U1A protein with F37M and F77M mutations for improved phasing | | Descriptor: | ACETATE ION, BETA-MERCAPTOETHANOL, SODIUM ION, ... | | Authors: | Jenkins, J.L, Lippa, G.M, Wedekind, J.E. | | Deposit date: | 2021-01-26 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Affinity and Structural Analysis of the U1A RNA Recognition Motif with Engineered Methionines to Improve Experimental Phasing
Crystals, 11, 2021
|
|
7A8W
 
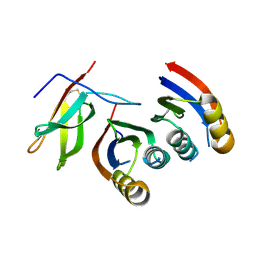 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikC with an engineered HMA domain of Pikp-1 from rice (Oryza sativa) | | Descriptor: | NBS-LRR class disease resistance protein, Uncharacterized protein | | Authors: | Maidment, J.H.R, De la Concepcion, J.C, Franceschetti, M, Banfield, M.J. | | Deposit date: | 2020-08-31 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The allelic rice immune receptor Pikh confers extended resistance to strains of the blast fungus through a single polymorphism in the effector binding interface.
Plos Pathog., 17, 2021
|
|
6IDN
 
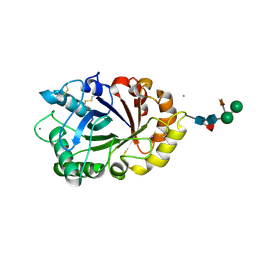 | | Crystal structure of ICChI chitinase from ipomoea carnea | | Descriptor: | CALCIUM ION, ICChI, a glycosylated chitinase, ... | | Authors: | Kumar, S, Kumar, A, Patel, A.K. | | Deposit date: | 2018-09-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | TIM barrel fold and glycan moieties in the structure of ICChI, a protein with chitinase and lysozyme activity.
Phytochemistry, 170, 2020
|
|
6BIE
 
 | |
7Q7K
 
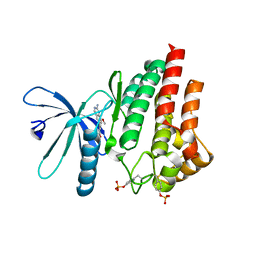 | | JAK2 in complex with 4-(2-amino-8-methoxyquinazolin-6-yl)phenol | | Descriptor: | 4-(2-azanyl-8-methoxy-quinazolin-6-yl)phenol, Tyrosine-protein kinase JAK2 | | Authors: | Rowland, P. | | Deposit date: | 2021-11-09 | | Release date: | 2022-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Investigation of Janus Kinase (JAK) Inhibitors for Lung Delivery and the Importance of Aldehyde Oxidase Metabolism.
J.Med.Chem., 65, 2022
|
|
7ADU
 
 | |
