4OED
 
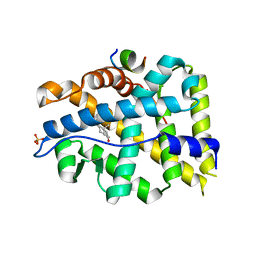 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, Protein BUD31 homolog, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-13 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
4OEZ
 
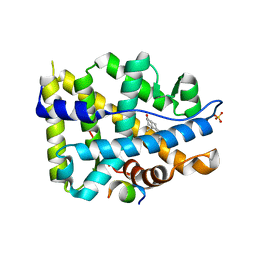 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, SULFATE ION, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-14 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
2QMJ
 
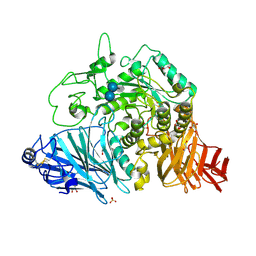 | | Crystral Structure of the N-terminal Subunit of Human Maltase-Glucoamylase in Complex with Acarbose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2007-07-16 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Human intestinal maltase-glucoamylase: crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity
J.Mol.Biol., 375, 2008
|
|
4OEY
 
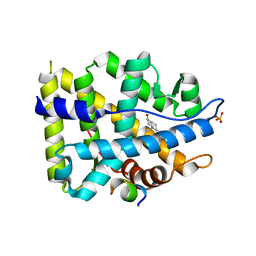 | | Crystal structure of AR-LBD bound with co-regulator peptide | | Descriptor: | 5-ALPHA-DIHYDROTESTOSTERONE, Androgen receptor, SULFATE ION, ... | | Authors: | Liu, J.S, Hsu, C.L, Wu, W.G. | | Deposit date: | 2014-01-14 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Identification of a new androgen receptor (AR) co-regulator BUD31 and related peptides to suppress wild-type and mutated AR-mediated prostate cancer growth via peptide screening and X-ray structure analysis.
Mol Oncol, 8, 2014
|
|
2QLY
 
 | | Crystral Structure of the N-terminal Subunit of Human Maltase-Glucoamylase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Sim, L, Rose, D.R. | | Deposit date: | 2007-07-13 | | Release date: | 2008-01-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Human intestinal maltase-glucoamylase: crystal structure of the N-terminal catalytic subunit and basis of inhibition and substrate specificity
J.Mol.Biol., 375, 2008
|
|
2KVH
 
 | |
2KVF
 
 | |
2KVG
 
 | |
3UZY
 
 | | Crystal structure of 5beta-reductase (AKR1D1) E120H mutant in complex with NADP+ and 5beta-dihydrotestosterone | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, 5-beta-DIHYDROTESTOSTERONE, CHLORIDE ION, ... | | Authors: | Chen, M, Christianson, D.W, Penning, T.M. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.832 Å) | | Cite: | Conversion of Human Steroid 5beta-Reductase (AKR1D1) into 3β-Hydroxysteroid Dehydrogenase by Single Point Mutation E120H: EXAMPLE OF PERFECT ENZYME ENGINEERING.
J.Biol.Chem., 287, 2012
|
|
4ITD
 
 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*(C6G)P*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | Deposit date: | 2013-01-18 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
4J6D
 
 | |
4IJ0
 
 | | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing transition mutation | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*(C6G)P*AP*AP*TP*TP*CP*GP*CP*G)-3'), STRONTIUM ION | | Authors: | Zhang, F, Suzuki, K, Tsunoda, M, Wilkinson, O, Millington, C.L, Williams, D.M, Morishita, E.C, Takenaka, A. | | Deposit date: | 2012-12-20 | | Release date: | 2013-05-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structures of DNA duplexes containing O6-carboxymethylguanine, a lesion associated with gastrointestinal cancer, reveal a mechanism for inducing pyrimidine transition mutations
Nucleic Acids Res., 41, 2013
|
|
3UNL
 
 | | Crystal structure of ketosteroid isomerase F54G from Pseudomonas testosteroni | | Descriptor: | SULFATE ION, Steroid Delta-isomerase | | Authors: | Gonzalez, A, Tsai, Y, Schwans, J, Sunden, F, Herschlag, D. | | Deposit date: | 2011-11-15 | | Release date: | 2012-11-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Crystal structure of ketosteroid isomerase F54G from Pseudomonas testosteroni
To be Published
|
|
2FW2
 
 | | Catalytic domain of CDY | | Descriptor: | Testis-specific chromodomain protein Y 2 | | Authors: | Min, J.R, Antoshenko, T, Wu, H, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A.M, Bochkarev, A, Plotnikov, A.N, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-01-31 | | Release date: | 2006-02-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of catalytic domain of Human Testis-specific chromodomain protein Y.
To be Published
|
|
7MZQ
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with fucose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, beta-L-fucopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZO
 
 | | Crystal structure of the UcaD lectin-binding domain | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZS
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with galactose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, alpha-D-galactopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZP
 
 | | Crystal structure of the UclD lectin-binding domain | | Descriptor: | F17-like fimbril adhesin subunit UclD, IODIDE ION | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZR
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with glucose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, beta-D-glucopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
6CDY
 
 | | Crystal structure of TEAD complexed with its inhibitor | | Descriptor: | 2-[(4H-1,2,4-triazol-3-yl)sulfanyl]-N-{4-[(3s,5s,7s)-tricyclo[3.3.1.1~3,7~]decan-1-yl]phenyl}acetamide, Transcriptional enhancer factor TEF-4 | | Authors: | LIU, S, HAN, X, LUO, X. | | Deposit date: | 2018-02-09 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Lats1/2 Sustain Intestinal Stem Cells and Wnt Activation through TEAD-Dependent and Independent Transcription.
Cell Stem Cell, 26, 2020
|
|
1DC9
 
 | | PROPERTIES AND CRYSTAL STRUCTURE OF A BETA-BARREL FOLDING MUTANT, V60N INTESTINAL FATTY ACID BINDING PROTEIN (IFABP) | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN | | Authors: | Ropson, I.J, Yowler, B.C, Dalessio, P.M, Banaszak, L, Thompson, J. | | Deposit date: | 1999-11-04 | | Release date: | 2000-03-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Properties and crystal structure of a beta-barrel folding mutant.
Biophys.J., 78, 2000
|
|
6KLA
 
 | | Crystal structure of human c-KIT kinase domain in complex with compound 15a | | Descriptor: | Mast/stem cell growth factor receptor Kit, N-[6-(4-ethylpiperazin-1-yl)-2-methyl-pyrimidin-4-yl]-5-pyridin-4-yl-1,3-thiazol-2-amine | | Authors: | Wu, T.S, Peng, Y.H, Hsueh, C.C, Wu, S.Y. | | Deposit date: | 2019-07-30 | | Release date: | 2019-11-27 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.109 Å) | | Cite: | Identification of a Multitargeted Tyrosine Kinase Inhibitor for the Treatment of Gastrointestinal Stromal Tumors and Acute Myeloid Leukemia.
J.Med.Chem., 62, 2019
|
|
6GSR
 
 | | Single Particle Cryo-EM map of human Transferrin receptor 1 - H-Ferritin complex at 5.5 Angstrom resolution. | | Descriptor: | Ferritin heavy chain, Transferrin receptor protein 1 | | Authors: | Testi, C, Montemiglio, L.C, Vallone, B, Des Georges, A, Boffi, A, Mancia, F, Baiocco, P. | | Deposit date: | 2018-06-15 | | Release date: | 2019-03-27 | | Last modified: | 2019-05-08 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Cryo-EM structure of the human ferritin-transferrin receptor 1 complex.
Nat Commun, 10, 2019
|
|
6H5I
 
 | | Single Particle Cryo-EM map of human Transferrin receptor 1 - H-Ferritin complex. | | Descriptor: | Ferritin heavy chain, Transferrin receptor protein 1 | | Authors: | Testi, C, Montemiglio, L.C, Vallone, B, Des Georges, A, Boffi, A, Mancia, F, Baiocco, P, Savino, C. | | Deposit date: | 2018-07-24 | | Release date: | 2019-03-27 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of the human ferritin-transferrin receptor 1 complex.
Nat Commun, 10, 2019
|
|
1IMT
 
 | | MAMBA INTESTINAL TOXIN 1, NMR, 39 STRUCTURES | | Descriptor: | INTESTINAL TOXIN 1 | | Authors: | Boisbouvier, J, Albrand, J.-P, Blackledge, M, Jaquinod, M, Schweitz, H, Lazdunski, M, Marion, D. | | Deposit date: | 1998-04-14 | | Release date: | 1999-04-20 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | A structural homologue of colipase in black mamba venom revealed by NMR floating disulphide bridge analysis.
J.Mol.Biol., 283, 1998
|
|
