7OSW
 
 | |
7PEQ
 
 | | Model of the outer rings of the human nuclear pore complex | | Descriptor: | Nuclear pore complex protein Nup107, Nuclear pore complex protein Nup133, Nuclear pore complex protein Nup160, ... | | Authors: | Schuller, A.P, Wojtynek, M, Mankus, D, Tatli, M, Kronenberg-Tenga, R, Regmi, S.G, Dasso, M, Weis, K, Medalia, O, Schwartz, T.U. | | Deposit date: | 2021-08-11 | | Release date: | 2021-10-20 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (35 Å) | | Cite: | The cellular environment shapes the nuclear pore complex architecture.
Nature, 598, 2021
|
|
7Q4O
 
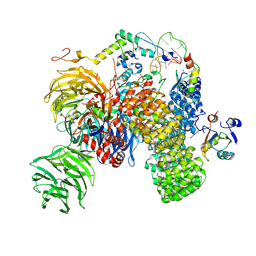 | | Substrate-bound A-like U2 snRNP | | Descriptor: | BPS oligo, PHD finger-like domain-containing protein 5A, Splicing factor 3A subunit 2, ... | | Authors: | Tholen, J, Galej, W.P. | | Deposit date: | 2021-11-01 | | Release date: | 2022-03-30 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural basis of branch site recognition by the human spliceosome.
Science, 375, 2022
|
|
7Q4P
 
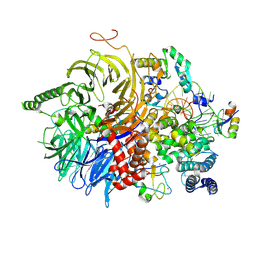 | | U2 snRNP after ATP-dependent remodelling | | Descriptor: | PHD finger-like domain-containing protein 5A, Splicing factor 3A subunit 2, Splicing factor 3A subunit 3, ... | | Authors: | Tholen, J, Galej, W.P. | | Deposit date: | 2021-11-01 | | Release date: | 2022-03-30 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.15 Å) | | Cite: | Structural basis of branch site recognition by the human spliceosome.
Science, 375, 2022
|
|
6SZS
 
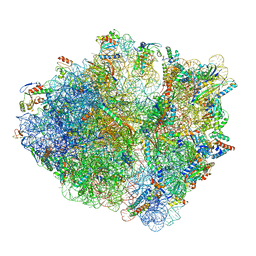 | | Release factor-dependent ribosome rescue by BrfA in the Gram-positive bacterium Bacillus subtilis | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Muller, C, Beckert, B, Wilson, D.N. | | Deposit date: | 2019-10-02 | | Release date: | 2019-12-04 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Release factor-dependent ribosome rescue by BrfA in the Gram-positive bacterium Bacillus subtilis.
Nat Commun, 10, 2019
|
|
6BY1
 
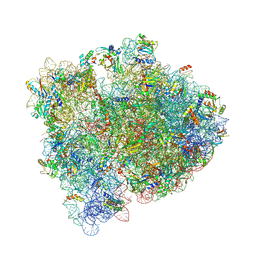 | | E. coli pH03H9 complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Amiri, H, Noller, H.F. | | Deposit date: | 2017-12-19 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.94 Å) | | Cite: | Structural evidence for product stabilization by the ribosomal mRNA helicase.
Rna, 25, 2019
|
|
7S1K
 
 | | Cfr-modified Escherichia coli stalled ribosome with antibiotic radezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Tsai, K, Stojkovic, V, Lee, D.J, Young, I.D, Szal, T, Vazquez-Laslop, N, Mankin, A.S, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2022-01-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SSW
 
 | |
7SSN
 
 | |
7SS9
 
 | | Late translocation intermediate with EF-G partially dissociated (Structure V) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Carbone, C.E, Loveland, A.B, Gamper, H.B, Hou, Y, Korostelev, A.A. | | Deposit date: | 2021-11-10 | | Release date: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Time-resolved cryo-EM visualizes ribosomal translocation with EF-G and GTP.
Nat Commun, 12, 2021
|
|
7ST7
 
 | |
7ST6
 
 | |
7SSL
 
 | |
7ST2
 
 | |
7QCD
 
 | |
7S1I
 
 | | Wild-type Escherichia coli stalled ribosome with antibiotic radezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7S1J
 
 | | Wild-type Escherichia coli ribosome with antibiotic radezolid | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Young, I.D, Stojkovic, V, Tsai, K, Lee, D.J, Fraser, J.S, Galonic Fujimori, D. | | Deposit date: | 2021-09-02 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structural basis for context-specific inhibition of translation by oxazolidinone antibiotics.
Nat.Struct.Mol.Biol., 29, 2022
|
|
8PVA
 
 | | Structure of bacterial ribosome determined by cryoEM at 100 keV | | Descriptor: | 16S rRNA, 23S rRNA, 50S ribosomal protein L14, ... | | Authors: | McMullan, G, Naydenova, K, Mihaylov, D, Peet, M.J, Wilson, H, Yamashita, K, Dickerson, J.L, Chen, S, Cannone, G, Lee, Y, Hutchings, K.A, Gittins, O, Sobhy, M, Wells, T, El-Gomati, M.M, Dalby, J, Meffert, M, Schulze-Briese, C, Henderson, R, Russo, C.J. | | Deposit date: | 2023-07-17 | | Release date: | 2023-11-29 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure determination by cryoEM at 100 keV.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7SSO
 
 | |
7SSD
 
 | |
8PHJ
 
 | | cA4-bound Cami1 in complex with 70S ribosome | | Descriptor: | 16S rRNA, 23S rRNA (2862-MER), 5S rRNA, ... | | Authors: | Tamulaitiene, G, Mogila, I, Sasnauskas, G, Tamulaitis, G. | | Deposit date: | 2023-06-20 | | Release date: | 2023-12-13 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.67 Å) | | Cite: | Ribosomal stalk-captured CARF-RelE ribonuclease inhibits translation following CRISPR signaling.
Science, 382, 2023
|
|
6TE1
 
 | | Structure of the KDM1A/CoREST complex with the inhibitor 2-[3-{4-chloro-3-[(4-chlorophenyl)ethynyl]phenyl}-1-(3-morpholin-4-ylpropyl)-1,4,6,7-tetrahydro-5H-pyrazolo[4,3-c]pyridin-5-yl]-2-oxoethanol | | Descriptor: | 5-[4-cyclobutyl-1-[2-(4-piperidin-4-yloxyphenoxy)ethyl]imidazol-2-yl]-4-methyl-thieno[3,2-b]pyrrole, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Pasqualato, S, Cecatiello, V. | | Deposit date: | 2019-11-11 | | Release date: | 2020-06-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Discovery of Reversible Inhibitors of KDM1A Efficacious in Acute Myeloid Leukemia Models.
Acs Med.Chem.Lett., 11, 2020
|
|
8P18
 
 | | E167K RF2 on E. coli 70S release complex with UGG (Structure III) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Pundir, S, Larsson, D.S.D, Selmer, M, Sanyal, S. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | The compensatory mechanism of a naturally evolved
E167K RF2 counteracting the loss of
RF1 in bacteria
To Be Published
|
|
8P16
 
 | | E167K RF2 on E. coli 70S release complex with UGG (Structure I) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S rRNA, 23S rRNA, ... | | Authors: | Pundir, S, Larsson, D.S.D, Selmer, M, Sanyal, S. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | The compensatory mechanism of a naturally evolved
E167K RF2 counteracting the loss of
RF1 in bacteria
To Be Published
|
|
8P17
 
 | | E167K RF2 on E. coli 70S release complex with UGG (Structure II) | | Descriptor: | 1,4-DIAMINOBUTANE, 16S RNA, 23S rRNA, ... | | Authors: | Pundir, S, Larsson, D.S.D, Selmer, M, Sanyal, S. | | Deposit date: | 2023-05-11 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | The compensatory mechanism of a naturally evolved
E167K RF2 counteracting the loss of
RF1 in bacteria
To Be Published
|
|
