5GYK
 
 | | Crystal Structure of Mdm12-deletion mutant | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Mitochondrial distribution and morphology protein 12 | | Authors: | Jeong, H, Park, J, Lee, C. | | Deposit date: | 2016-09-22 | | Release date: | 2016-11-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.596 Å) | | Cite: | Crystal structure of Mdm12 reveals the architecture and dynamic organization of the ERMES complex
EMBO Rep., 17, 2016
|
|
7QU7
 
 | | ATAD2 in complex with FragLite16 | | Descriptor: | 1,2-ETHANEDIOL, 4-bromanyl-1,8-naphthyridine, ATPase family AAA domain-containing protein 2, ... | | Authors: | Turberville, S, Martin, M.P, Hope, I, Noble, M.E.M. | | Deposit date: | 2022-01-17 | | Release date: | 2022-11-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Mapping Ligand Interactions of Bromodomains BRD4 and ATAD2 with FragLites and PepLites─Halogenated Probes of Druglike and Peptide-like Molecular Interactions.
J.Med.Chem., 65, 2022
|
|
7PEB
 
 | |
7PEA
 
 | |
7PEC
 
 | |
6YGA
 
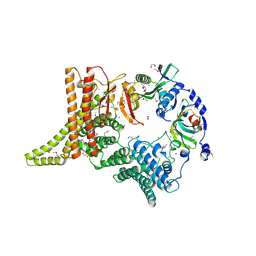 | | Crystal structure of the apo NatC complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Grunwald, S, Hopf, L, Bock-Bierbaum, T, Lally, C.C, Spahn, C.M.T, Daumke, O. | | Deposit date: | 2020-03-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Divergent architecture of the heterotrimeric NatC complex explains N-terminal acetylation of cognate substrates.
Nat Commun, 11, 2020
|
|
4WMW
 
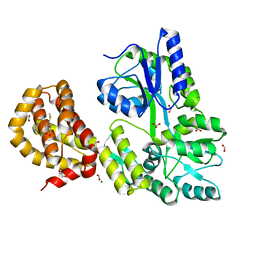 | | The structure of MBP-MCL1 bound to ligand 5 at 1.9A | | Descriptor: | 1,2-ETHANEDIOL, 2-hydroxy-5-(methylsulfanyl)benzoic acid, FORMIC ACID, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
6HCN
 
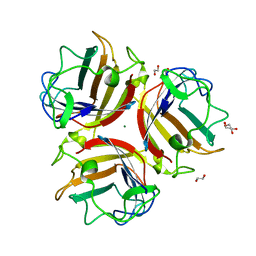 | | Adenovirus Type 5 Fiber Knob protein at 1.49A resolution | | Descriptor: | 1,2-ETHANEDIOL, Fiber protein, MAGNESIUM ION | | Authors: | Rizkallah, P.J, Parker, A.L, Baker, A.T. | | Deposit date: | 2018-08-15 | | Release date: | 2019-02-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Diversity within the adenovirus fiber knob hypervariable loops influences primary receptor interactions.
Nat Commun, 10, 2019
|
|
8RHQ
 
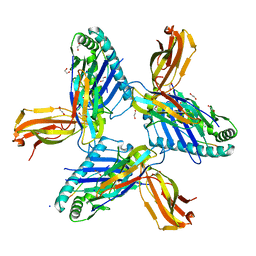 | | Crystal structure of HLA-A*11:01 in complex with SVLNDIFSRL, an 10-mer epitope from SARS-CoV-2 Spike (S975-984) | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CHLORIDE ION, ... | | Authors: | Ahn, Y.M, Maddumage, J.C, Szeto, C, Gras, S. | | Deposit date: | 2023-12-16 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The impact of SARS-CoV-2 spike mutation on peptide presentation is HLA allomorph-specific.
Curr Res Struct Biol, 7, 2024
|
|
6PYU
 
 | | Human PI3Kdelta in complex with Compound 4-2 ((3S)-1'-(cyclopropanecarbonyl)-5-(quinoxalin-6-yl)spiro[indole-3,2'-pyrrolidin]-2(1H)-one) | | Descriptor: | (3S)-1'-(cyclopropanecarbonyl)-5-(quinoxalin-6-yl)spiro[indole-3,2'-pyrrolidin]-2(1H)-one, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit delta isoform | | Authors: | Lesburg, C.A, Augustin, M.A. | | Deposit date: | 2019-07-30 | | Release date: | 2019-08-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Design of selective PI3K delta inhibitors using an iterative scaffold-hopping workflow.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6W5L
 
 | | 2.1 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7g | | Descriptor: | (2~{S})-~{N}-[(1~{R})-1-[bis($l^{1}-oxidanyl)-methoxy-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]-2-[[[2-(3-chlorophenyl)-2-methyl-propoxy]-oxidanylidene-methyl]amino]-4-methyl-pentanamide, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
5MZ8
 
 | | Crystal structure of aldehyde dehydrogenase 21 (ALDH21) from Physcomitrella patens in complex with the reaction product succinate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Kopecny, D, Vigouroux, A, Briozzo, P, Morera, S. | | Deposit date: | 2017-01-31 | | Release date: | 2017-08-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The ALDH21 gene found in lower plants and some vascular plants codes for a NADP(+) -dependent succinic semialdehyde dehydrogenase.
Plant J., 92, 2017
|
|
6VW9
 
 | | C-terminal regulatory domain of the chloride transporter KCC-1 from C. elegans, proteolyzed during crystallization | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, GLYCEROL, K+/Cl-Cotransporter | | Authors: | Zimanyi, C.M, Cheung, J. | | Deposit date: | 2020-02-19 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the Regulatory Cytosolic Domain of a Eukaryotic Potassium-Chloride Cotransporter.
Structure, 28, 2020
|
|
4WMS
 
 | | STRUCTURE OF APO MBP-MCL1 AT 1.9A | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, MAGNESIUM ION, ... | | Authors: | Clifton, M.C, Dranow, D.M. | | Deposit date: | 2014-10-09 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Maltose-Binding Protein Fusion Construct Yields a Robust Crystallography Platform for MCL1.
Plos One, 10, 2015
|
|
8RPH
 
 | | JanthE from Janthinobacterium sp. HH01,ketobutyryl-ThDP | | Descriptor: | (2~{S})-2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-4-methyl-5-[2-[oxidanyl(phosphonooxy)phosphoryl]oxyethyl]-1,3-thiazol-2-yl]-2-oxidanyl-butanoic acid, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Lanza, L, Leogrande, C, Rabe von Pappenheim, F, Tittmann, K, Mueller, M. | | Deposit date: | 2024-01-16 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Identification and Characterization of Thiamine Diphosphate-Dependent Lyases with an Unusual CDG Motif.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8RCP
 
 | | Structure of Human Serum Albumin in complex with Myristic Acid | | Descriptor: | 1,2-ETHANEDIOL, MYRISTIC ACID, Serum albumin | | Authors: | Pomyalov, S, Sidorenko, V.S, Grollman, A.P, Shoham, G. | | Deposit date: | 2023-12-06 | | Release date: | 2024-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic insights into the transport of aristolochic acids and their active metabolites by human serum albumin.
J.Biol.Chem., 300, 2024
|
|
6HOY
 
 | |
4WX2
 
 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with two F6F molecules in the alpha-site and one F6F molecule in the beta-site | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Caulkins, B.G, Young, R.P, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2014-11-13 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Visualizing the tunnel in tryptophan synthase with crystallography: Insights into a selective filter for accommodating indole and rejecting water.
Biochim.Biophys.Acta, 1864, 2016
|
|
6HLY
 
 | | Structure in P212121 form of the PBP AgtB in complex with agropinic acid from A.tumefacien R10 | | Descriptor: | 1,2-ETHANEDIOL, Agropine permease, agropinic acid | | Authors: | Morera, S, Marty, L, Vigouroux, A. | | Deposit date: | 2018-09-11 | | Release date: | 2018-12-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for two efficient modes of agropinic acid opine import into the bacterial pathogenAgrobacterium tumefaciens.
Biochem. J., 476, 2019
|
|
9BP2
 
 | |
8VSM
 
 | | Ternary structure of 14-3-3 sigma, ARAF phosphopeptide (pS214) and compound 78 (1124378) | | Descriptor: | 1-[8-(4-bromophenyl)sulfonyl-5-oxa-2,8-diazaspiro[3.5]nonan-2-yl]-2-chloranyl-ethanone, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Vickery, H.R, Virta, J.M, Pennings, M, Konstantinidou, M, van den Oetelaar, M, Neitz, R.J, Ottmann, C, Brunsveld, L, Arkin, M.R. | | Deposit date: | 2024-01-24 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ternary structure of 14-3-3 sigma, ARAF phosphopeptide (pS214) and compound 78 (1124378)
To Be Published
|
|
6HNJ
 
 | | The ligand-bound, open structure of CD0873, a substrate binding protein with adhesive properties from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, ABC-type transport system, sugar-family extracellular solute-binding protein, ... | | Authors: | Bradshaw, W.J, Kovacs-Simon, A, Harmer, N.J, Michell, S.L, Acharya, K.R. | | Deposit date: | 2018-09-15 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Molecular features of lipoprotein CD0873: A potential vaccine against the human pathogenClostridioides difficile.
J.Biol.Chem., 294, 2019
|
|
4X6M
 
 | |
7P1A
 
 | | Carbonic Anhydrase VII Sultam Based Inhibitors | | Descriptor: | 4-[2-[4-[(4-methylphenyl)methyl]-1,1-bis(oxidanylidene)-1,2,4-thiadiazinan-2-yl]ethyl]benzenesulfonamide, Carbonic anhydrase 7, GLYCEROL, ... | | Authors: | D'Ambrosio, K, De Simone, G. | | Deposit date: | 2021-07-01 | | Release date: | 2021-11-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Sultam based Carbonic Anhydrase VII inhibitors for the management of neuropathic pain.
Eur.J.Med.Chem., 227, 2021
|
|
4WJ1
 
 | |
