3EXB
 
 | |
4MSH
 
 | | Crystal Structure of PDE10A2 with fragment ZT0143 ((2S)-4-chloro-2,3-dihydro-1,3-benzothiazol-2-amine) | | Descriptor: | 4-chloro-1,3-benzothiazol-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MO1
 
 | | Crystal structure of antitermination protein Q from bacteriophage lambda. Northeast Structural Genomics Consortium target OR18A. | | Descriptor: | Antitermination protein Q, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Vorobiev, S, Su, M, Nickels, B, Seetharaman, J, Sahdev, S, Xiao, R, Kogan, S, Maglaqui, M, Wang, D, Everett, J.K, Acton, T.B, Ebright, R.H, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-09-11 | | Release date: | 2013-09-25 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal structure of antitermination protein Q from bacteriophage lambda.
To be Published
|
|
4MRW
 
 | | Crystal structure of PDE10A2 with fragment ZT0120 (7-chloroquinolin-4-ol) | | Descriptor: | 7-chloroquinolin-4-ol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, NIenaber, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MSE
 
 | | Crystal structure of PDE10A2 with fragment ZT1597 (2-({[(2S)-2-methyl-2,3-dihydro-1,3-benzothiazol-5-yl]oxy}methyl)quinoline) | | Descriptor: | 2-{[(2-methyl-1,3-benzothiazol-5-yl)oxy]methyl}quinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MRZ
 
 | | Crystal structure of PDE10A2 with fragment ZT0429 (4-methyl-3-nitropyridin-2-amine) | | Descriptor: | 4-methyl-3-nitropyridin-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-17 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MSA
 
 | | Crystal structure of PDE10A2 with fragment ZT0449 (5-nitro-1H-benzimidazole) | | Descriptor: | 5-nitro-1H-benzimidazole, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MSC
 
 | | Crystal structure of PDE10A2 with fragment ZT1595 (2-[(quinolin-7-yloxy)methyl]quinoline) | | Descriptor: | 2-[(quinolin-7-yloxy)methyl]quinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MS0
 
 | | Crystal structure of PDE10A2 with fragment ZT0443 (6-chloropyrimidine-2,4-diamine) | | Descriptor: | 6-chloropyrimidine-2,4-diamine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
7W64
 
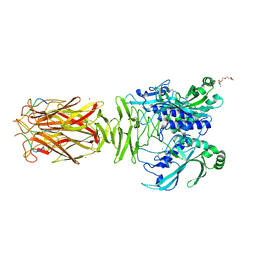 | | Crystal structure of minor pilin TcpB from Vibrio cholerae complexed with N-terminal peptide fragment of TcpF | | Descriptor: | CALCIUM ION, CHLORIDE ION, PENTAETHYLENE GLYCOL, ... | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
7W65
 
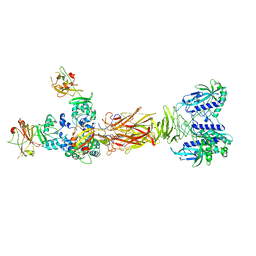 | | Crystal structure of minor pilin TcpB from Vibrio cholerae complexed with secreted protein TcpF | | Descriptor: | Toxin coregulated pilus biosynthesis protein F, Toxin-coregulated pilus biosynthesis protein B | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
7W63
 
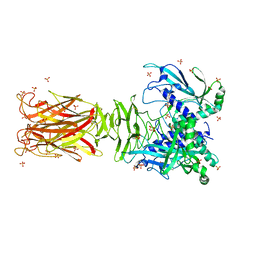 | | Crystal structure of minor pilin TcpB from Vibrio cholerae | | Descriptor: | SULFATE ION, Toxin-coregulated pilus biosynthesis protein B | | Authors: | Oki, H, Kawahara, K, Iimori, M, Imoto, Y, Maruno, T, Uchiyama, S, Muroga, Y, Yoshida, A, Yoshida, T, Ohkubo, T, Matsuda, S, Iida, T, Nakamura, S. | | Deposit date: | 2021-12-01 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-16 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structural basis for the toxin-coregulated pilus-dependent secretion of Vibrio cholerae colonization factor.
Sci Adv, 8, 2022
|
|
4L1L
 
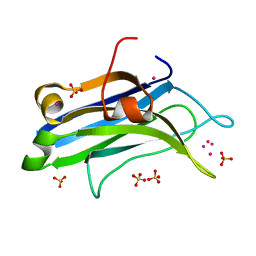 | | Rat PKC C2 domain bound to CD | | Descriptor: | CADMIUM ION, Protein kinase C alpha type, SULFATE ION | | Authors: | Morales, K.M, Yang, Y, Long, Z, Li, P, Taylor, A.B, Hart, P.J, Igumenova, T.I. | | Deposit date: | 2013-06-03 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cd(2+) as a ca(2+) surrogate in protein-membrane interactions: isostructural but not isofunctional.
J.Am.Chem.Soc., 135, 2013
|
|
4MZJ
 
 | | Crystal Structure of MTIP from Plasmodium falciparum in complex with pGly[801,805], a stapled myoA tail peptide | | Descriptor: | Myosin A tail domain interacting protein, Myosin-A | | Authors: | Douse, C.H, Garnett, J.A, Maas, S.J, Cota, E, Tate, E.W. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.474 Å) | | Cite: | Crystal Structures of Stapled and Hydrogen Bond Surrogate Peptides Targeting a Fully Buried Protein-Helix Interaction.
Acs Chem.Biol., 8, 2014
|
|
4LLJ
 
 | | Crystal structure of PDE10A2 with fragment ZT214 | | Descriptor: | 2H-isoindole-1,3-diamine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM0
 
 | | Crystal structure of PDE10A2 with fragment ZT448 | | Descriptor: | 5-NITROINDAZOLE, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LLK
 
 | | Crystal structure of PDE10A2 with fragment ZT217 | | Descriptor: | 2-methylquinazolin-4(3H)-one, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LKQ
 
 | | Crystal structure of PDE10A2 with fragment ZT017 | | Descriptor: | 4-amino-1,7-dihydro-6H-pyrazolo[3,4-d]pyrimidine-6-thione, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4MSN
 
 | | Crystal structure of PDE10A2 with fragment ZT0451 (8-nitroquinoline) | | Descriptor: | 8-nitroquinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-09-18 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Identification and optimization of PDE10A inhibitors using fragment-based screening by nanocalorimetry and X-ray crystallography.
J Biomol Screen, 19, 2014
|
|
4MZK
 
 | | Crystal Structure of MTIP from Plasmodium falciparum in complex with pGly[807,811], a stapled myoA tail peptide | | Descriptor: | Myosin A tail domain interacting protein, pGly[807,811], a stapled myoA tail peptide | | Authors: | Douse, C.H, Garnett, J.A, Maas, S.J, Cota, E, Tate, E.W. | | Deposit date: | 2013-09-30 | | Release date: | 2013-11-06 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structures of Stapled and Hydrogen Bond Surrogate Peptides Targeting a Fully Buried Protein-Helix Interaction.
Acs Chem.Biol., 8, 2014
|
|
4LM3
 
 | | Crystal structure of PDE10A2 with fragment ZT464 | | Descriptor: | 1,2-ETHANEDIOL, 1-(2,3-dihydro-1,4-benzodioxin-6-yl)ethanone, NICKEL (II) ION, ... | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM1
 
 | | Crystal structure of PDE10A2 with fragment ZT450 | | Descriptor: | 5-nitroquinoline, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LLX
 
 | | Crystal structure of PDE10A2 with fragment ZT434 | | Descriptor: | 4,6-dimethylpyrimidin-2-amine, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
4LM2
 
 | | Crystal structure of PDE10A2 with fragment ZT462 | | Descriptor: | 2,3-dihydro-1,4-benzodioxin-6-ylmethanol, NICKEL (II) ION, cAMP and cAMP-inhibited cGMP 3',5'-cyclic phosphodiesterase 10A | | Authors: | Sridhar, V, Badger, J, Logan, C, Chie-Leon, B, Nienaber, V. | | Deposit date: | 2013-07-09 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Optimization of PDE10A Inhibitors Using Fragment-Based Screening by Nanocalorimetry and X-ray Crystallography.
J Biomol Screen, 19, 2014
|
|
7X5G
 
 | | Nrf2 (A510Y)-MafG heterodimer bound with CsMBE2 | | Descriptor: | DNA (5'-D(*CP*AP*CP*AP*GP*TP*GP*AP*CP*TP*CP*AP*GP*CP*AP*G)-3'), DNA (5'-D(*GP*CP*TP*GP*CP*TP*GP*AP*GP*TP*CP*AP*CP*TP*GP*T)-3'), Nuclear factor erythroid 2-related factor 2, ... | | Authors: | Sengoku, T, Shiina, M, Suzuki, K, Hamada, K, Sato, K, Uchiyama, A, Okada, C, Baba, S, Ohta, T, Motohashi, H, Yamamoto, M, Ogata, K. | | Deposit date: | 2022-03-04 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of transcription regulation by CNC family transcription factor, Nrf2.
Nucleic Acids Res., 50, 2022
|
|
