6VCL
 
 | | Crystal structure of E.coli RppH-DapF in complex with pppGpp, Mg2+ and F- | | Descriptor: | CHLORIDE ION, Diaminopimelate epimerase, FLUORIDE ION, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
6VCR
 
 | | Crystal structure of E.coli RppH in complex with CTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, PYROPHOSPHATE, RNA pyrophosphohydrolase, ... | | Authors: | Gao, A, Vasilyev, N, Kaushik, A, Duan, W, Serganov, A. | | Deposit date: | 2019-12-21 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Principles of RNA and nucleotide discrimination by the RNA processing enzyme RppH.
Nucleic Acids Res., 48, 2020
|
|
4Y66
 
 | |
3TDL
 
 | | Structure of human serum albumin in complex with DAUDA | | Descriptor: | 11-({[5-(dimethylamino)naphthalen-1-yl]sulfonyl}amino)undecanoic acid, MYRISTIC ACID, Serum albumin | | Authors: | Wang, Y, Luo, Z, Shi, X, Wang, H, Nie, L. | | Deposit date: | 2011-08-11 | | Release date: | 2012-06-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A fluorescent fatty acid probe, DAUDA, selectively displaces two myristates bound in human serum albumin
Protein Sci., 20, 2011
|
|
5MUT
 
 | | Crystal structure of potent human Dihydroorotate Dehydrogenase inhibitors based on hydroxylated azole scaffolds | | Descriptor: | 2-methyl-5-oxidanyl-~{N}-[2,3,5,6-tetrakis(fluoranyl)-4-phenyl-phenyl]-1,2,3-triazole-4-carboxamide, ACETATE ION, CHLORIDE ION, ... | | Authors: | Goyal, P, Andersson, M, Moritzer, A.C, Sainas, S, Pippione, A.C, Boschi, D, Al-Kadaraghi, S, Lolli, M, Friemann, R. | | Deposit date: | 2017-01-14 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray structural studies of potent human dihydroorotate dehydrogenase inhibitors based on hydroxylated azole scaffolds.
Eur J Med Chem, 129, 2017
|
|
8JB1
 
 | |
6C83
 
 | | Structure of Aurora A (122-403) bound to inhibitory Monobody Mb2 and AMPPCP | | Descriptor: | Aurora kinase A, Mb2 Monobody, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Hoemberger, M, Kutter, S, Zorba, A, Nguyen, V, Shohei, A, Shohei, K, Kern, D. | | Deposit date: | 2018-01-24 | | Release date: | 2019-02-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Allosteric modulation of a human protein kinase with monobodies.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1H2U
 
 | | Structure of the human nuclear cap-binding-complex (CBC) in complex with a cap analogue m7GpppG | | Descriptor: | 20 KDA NUCLEAR CAP BINDING PROTEIN, 7N-METHYL-8-HYDROGUANOSINE-5'-MONOPHOSPHATE, 80 KDA NUCLEAR CAP BINDING PROTEIN, ... | | Authors: | Mazza, C, Segref, A, Mattaj, I.W, Cusack, S. | | Deposit date: | 2002-08-16 | | Release date: | 2002-10-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Large-Scale Induced Fit Recognition of an M(7)Gpppg CAP Analogue by the Human Nuclear CAP-Binding Complex
Embo J., 21, 2002
|
|
8PII
 
 | | VHH Z70 mutant 3 in interaction with PHF6 Tau peptide | | Descriptor: | Microtubule-associated protein tau, VHH Z70 Mutant 3 | | Authors: | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | Deposit date: | 2023-06-21 | | Release date: | 2023-07-05 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
6GPS
 
 | | CRYSTAL STRUCTURE OF CCR2A IN COMPLEX WITH MK-0812 | | Descriptor: | C-C chemokine receptor type 2,Rubredoxin,C-C chemokine receptor type 2, ZINC ION, [(3~{S},4~{S})-3-methoxyoxan-4-yl]-[(1~{R},3~{S})-3-propan-2-yl-3-[[3-(trifluoromethyl)-7,8-dihydro-5~{H}-1,6-naphthyridin-6-yl]carbonyl]cyclopentyl]azanium | | Authors: | Pautsch, A, Schnapp, G. | | Deposit date: | 2018-06-07 | | Release date: | 2019-01-02 | | Last modified: | 2019-03-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal Structure of CC Chemokine Receptor 2A in Complex with an Orthosteric Antagonist Provides Insights for the Design of Selective Antagonists.
Structure, 27, 2019
|
|
6VFZ
 
 | | Crystal Structure of Human Mitochondrial Isocitrate Dehydrogenase (IDH2) R140Q Mutant Homodimer in Complex with NADPH and AG-881 (Vorasidenib) Inhibitor. | | Descriptor: | 6-(6-chloropyridin-2-yl)-N2,N4-bis[(2R)-1,1,1-trifluoropropan-2-yl]-1,3,5-triazine-2,4-diamine, CALCIUM ION, Isocitrate dehydrogenase [NADP], ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-01-07 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Vorasidenib (AG-881): A First-in-Class, Brain-Penetrant Dual Inhibitor of Mutant IDH1 and 2 for Treatment of Glioma.
Acs Med.Chem.Lett., 11, 2020
|
|
5M95
 
 | | STAPHYLOCOCCUS CAPITIS DIVALENT METAL ION TRANSPORTER (DMT) IN COMPLEX WITH MANGANESE | | Descriptor: | CAMELID ANTIBODY FRAGMENT, NANOBODY, Divalent metal cation transporter MntH, ... | | Authors: | Ehrnstorfer, I.A, Geertsma, E.R, Pardon, E, Steyaert, J, Dutzler, R. | | Deposit date: | 2016-10-31 | | Release date: | 2016-11-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure Of A Slc11 (Nramp) Transporter Reveals The Basis For Transition-Metal Ion Transport.
Nat.Struct.Mol.Biol., 21, 2014
|
|
8BK7
 
 | |
4Y7P
 
 | | Structure of alkaline D-peptidase from Bacillus cereus | | Descriptor: | Alkaline D-peptidase, THIOCYANATE ION | | Authors: | Nakano, S, Okazaki, S, Ishitsubo, E, Kawahara, N, Komeda, H, Tokiwa, H, Asano, Y. | | Deposit date: | 2015-02-15 | | Release date: | 2015-10-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and computational analysis of peptide recognition mechanism of class-C type penicillin binding protein, alkaline D-peptidase from Bacillus cereus DF4-B
Sci Rep, 5, 2015
|
|
6M6L
 
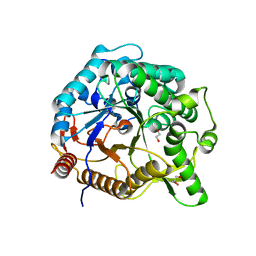 | | The crystal structure of glycosidase hydrolyzing Notoginsenoside | | Descriptor: | 1,2-ETHANEDIOL, Beta-glucosidase, GLYCEROL, ... | | Authors: | Wang, R.F. | | Deposit date: | 2020-03-15 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Characterization and Structural Elucidation of Enhanced Catalytic Activity upon Engineering Glycosidase KfGH01 for the Production of Vina-ginsenoside R7
To Be Published
|
|
8BD3
 
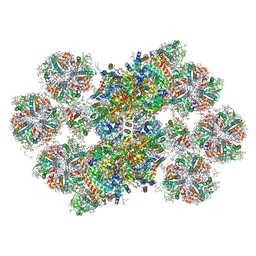 | | Cryo-EM structure of the Photosystem II - LHCII supercomplex from Chlorella ohadi | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R)-beta,beta-caroten-3-ol, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Fadeeva, M, Klaiman, D, Caspy, I, Nelson, N. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Structure of Chlorella ohadii Photosystem II Reveals Protective Mechanisms against Environmental Stress.
Cells, 12, 2023
|
|
5LMW
 
 | | Llama nanobody PorM_02 | | Descriptor: | GLYCEROL, Nanobody | | Authors: | Roche, J, Gaubert, A, Leone, P, Roussel, A. | | Deposit date: | 2016-08-01 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Camelid nanobodies used as crystallization chaperones for different constructs of PorM, a component of the type IX secretion system from Porphyromonas gingivalis.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
8BKG
 
 | |
4YKC
 
 | |
5LUT
 
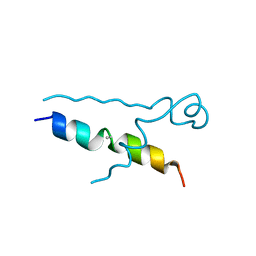 | | Structures of DHBN domain of Gallus gallus BLM helicase | | Descriptor: | BLM helicase, PHOSPHATE ION | | Authors: | Shi, J, Chen, W.-F, Zhang, B, Fan, S.-H, Ai, X, Liu, N.-N, Rety, S, Xi, X.-G. | | Deposit date: | 2016-09-09 | | Release date: | 2017-03-01 | | Last modified: | 2017-04-19 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | A helical bundle in the N-terminal domain of the BLM helicase mediates dimer and potentially hexamer formation.
J. Biol. Chem., 292, 2017
|
|
5LVU
 
 | | XiaF (apo) from Streptomyces sp. | | Descriptor: | SULFATE ION, XiaF protein | | Authors: | Kugel, S, Baunach, M, Baer, P, Ishida-Ito, M, Sundaram, S, Xu, Z, Groll, M, Hertweck, C. | | Deposit date: | 2016-09-14 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Cryptic indole hydroxylation by a non-canonical terpenoid cyclase parallels bacterial xenobiotic detoxification.
Nat Commun, 8, 2017
|
|
8BK8
 
 | |
6M6M
 
 | | The crystal structure of glycosidase mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucosidase | | Authors: | Wang, R.F. | | Deposit date: | 2020-03-15 | | Release date: | 2021-03-17 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Characterization and Structural Elucidation of Enhanced Catalytic Activity upon Engineering Glycosidase KfGH01 for the Production of Vina-ginsenoside R7
To Be Published
|
|
7M2N
 
 | | Crystal structure of Human Lactate Dehydrogenase A with Inhibitor Compound 15 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 5-[(5'-{1-(4-carboxy-1,3-thiazol-2-yl)-5-(cyclopropylmethyl)-4-[(3-fluoro-4-sulfamoylphenyl)methyl]-1H-pyrazol-3-yl}-2'-fluoro[1,1'-biphenyl]-4-yl)oxy]-1H-1,2,3-triazole-4-carboxylic acid, ... | | Authors: | Gumpena, R, Ding, J, Powell, D.A, Lowther, W.T. | | Deposit date: | 2021-03-17 | | Release date: | 2021-07-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Dual Glycolate Oxidase/Lactate Dehydrogenase A Inhibitors for Primary Hyperoxaluria
ACS Medicinal Chemistry Letters, 12, 2021
|
|
8PFU
 
 | | X-ray structure of the adduct formed upon reaction of Lysozyme with K3[Ru2(CO3)4] in condition A | | Descriptor: | 6,8-bis(4-fluorophenyl)-1,5-bis(oxidanyl)-2,4-dioxa-6,8-diaza-1$l^{4},5$l^{4}-diruthenabicyclo[3.3.0]octan-3-one, CARBONATE ION, Lysozyme C, ... | | Authors: | Teran, A, Ferraro, G, Merlino, A. | | Deposit date: | 2023-06-16 | | Release date: | 2023-07-19 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Charge effect in protein metalation reactions by diruthenium complexes
Inorg Chem Front, 2023
|
|
