2HBE
 
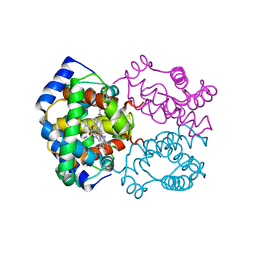 | | HIGH RESOLUTION X-RAY STRUCTURES OF MYOGLOBIN-AND HEMOGLOBIN-ALKYL ISOCYANIDE COMPLEXES | | Descriptor: | HEMOGLOBIN A (N-BUTYL ISOCYANIDE) (ALPHA CHAIN), HEMOGLOBIN A (N-BUTYL ISOCYANIDE) (BETA CHAIN), N-BUTYL ISOCYANIDE, ... | | Authors: | Johnson, K.A, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1994-08-31 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High Resolution X-Ray Structures of Myoglobin-and Hemoglobin-Alkyl Isocyanide Complexes
Thesis, 1, 1993
|
|
1RH9
 
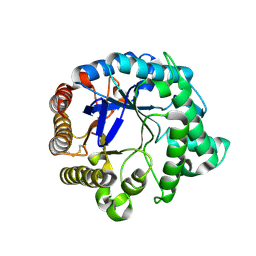 | | Family GH5 endo-beta-mannanase from Lycopersicon esculentum (tomato) | | Descriptor: | endo-beta-mannanase | | Authors: | Oakley, A.J, Bourgault, R, Bewley, J.D, Wilce, M.C.J. | | Deposit date: | 2003-11-14 | | Release date: | 2005-04-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-dimensional structure of (1,4)-beta-D-mannan mannanohydrolase from tomato fruit.
Protein Sci., 14, 2005
|
|
1RP9
 
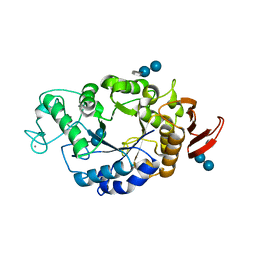 | | Crystal structure of barley alpha-amylase isozyme 1 (amy1) inactive mutant d180a in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-4-{[(1S,5R,6S)-3-formyl-5,6-dihydroxy-4-oxocyclohex-2-en-1-yl]amino}-alpha-D-xylo-hex-5-enopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase type 1 isozyme, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-12-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Binding to Barley {alpha}-Amylase 1
J.Biol.Chem., 280, 2005
|
|
2HF3
 
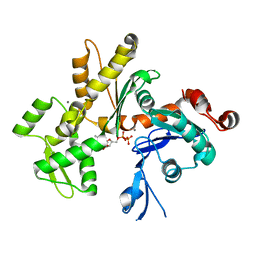 | | Crystal structure of monomeric Actin in the ADP bound state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-5C, CALCIUM ION | | Authors: | Rould, M.A, Wan, Q, Joel, P.B, Lowey, S, Trybus, K.M. | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structures of Expressed Non-polymerizable Monomeric Actin in the ADP and ATP States.
J.Biol.Chem., 281, 2006
|
|
2HFE
 
 | | Rb+ complex of a K channel with an amide to ester substitution in the selectivity filter | | Descriptor: | (2S)-2-(BUTYRYLOXY)-3-HYDROXYPROPYL NONANOATE, FAB Heavy Chain, FAB Light Chain, ... | | Authors: | Valiyaveetil, F.I, MacKinnon, R, Muir, T.W. | | Deposit date: | 2006-06-23 | | Release date: | 2006-09-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and Functional Consequences of an Amide-to-Ester Substitution in the Selectivity Filter of a Potassium Channel.
J.Am.Chem.Soc., 128, 2006
|
|
2HBF
 
 | | HIGH RESOLUTION X-RAY STRUCTURES OF MYOGLOBIN-AND HEMOGLOBIN-ALKYL ISOCYANIDE COMPLEXES | | Descriptor: | HEMOGLOBIN A (N-PROPYL ISOCYANIDE) (ALPHA CHAIN), HEMOGLOBIN A (N-PROPYL ISOCYANIDE) (BETA CHAIN), N-PROPYL ISOCYANIDE, ... | | Authors: | Johnson, K.A, Olson, J.S, Phillips Jr, G.N. | | Deposit date: | 1994-08-31 | | Release date: | 1995-02-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | High Resolution X-Ray Structures of Myoglobin-and Hemoglobin-Alkyl Isocyanide Complexes
Thesis, 1, 1993
|
|
1OKN
 
 | | CARBONIC ANHYDRASE II COMPLEX WITH THE 1OKN INHIBITOR 4-SULFONAMIDE-[1-(4-N-(5-FLUORESCEIN THIOUREA)BUTANE)] | | Descriptor: | 4-SULFONAMIDE-[4-(THIOMETHYLAMINOBUTANE)]BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Elbaum, D, Nair, S.K, Patchan, M.W, Thompson, R.B, Christianson, D.W. | | Deposit date: | 1996-06-25 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of a sulfonamide probe for fluorescence anisotropy detection of zinc with a carbonic anhydrase-based biosensor.
J.Am.Chem.Soc., 118, 1996
|
|
1OFV
 
 | | FLAVODOXIN FROM ANACYSTIS NIDULANS: REFINEMENT OF TWO FORMS OF THE OXIDIZED PROTEIN | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Smith, W.W, Pattridge, K.A, Luschinsky, C.L, Ludwig, M.L. | | Deposit date: | 1992-06-22 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined structures of oxidized flavodoxin from Anacystis nidulans.
J.Mol.Biol., 294, 1999
|
|
1R2H
 
 | | Human Bcl-XL containing an Ala to Leu mutation at position 142 | | Descriptor: | Apoptosis regulator Bcl-X | | Authors: | O'Neill, J.W, Manion, M.K, Giedt, C.D, Kim, K.M, Zhang, K.Y, Hockenbery, D.M. | | Deposit date: | 2003-09-26 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bcl-XL mutations suppress cellular sensitivity to antimycin A.
J.Biol.Chem., 279, 2004
|
|
1NMB
 
 | | THE STRUCTURE OF A COMPLEX BETWEEN THE NC10 ANTIBODY AND INFLUENZA VIRUS NEURAMINIDASE AND COMPARISON WITH THE OVERLAPPING BINDING SITE OF THE NC41 ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FAB NC10, ... | | Authors: | Malby, R.L, Tulip, W.R, Colman, P.M. | | Deposit date: | 1995-01-17 | | Release date: | 1995-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a complex between the NC10 antibody and influenza virus neuraminidase and comparison with the overlapping binding site of the NC41 antibody
Structure, 2, 1994
|
|
2HBM
 
 | | Structure of the yeast nuclear exosome component, Rrp6p, reveals an interplay between the active site and the HRDC domain; Protein in complex with Mn, Zn, and UMP | | Descriptor: | Exosome complex exonuclease RRP6, MANGANESE (II) ION, URIDINE-5'-MONOPHOSPHATE, ... | | Authors: | Midtgaard, S.F, Assenholt, J, Jonstrup, A.T, Van, L.B, Jensen, T.H, Brodersen, D.E. | | Deposit date: | 2006-06-14 | | Release date: | 2006-07-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the nuclear exosome component Rrp6p reveals an interplay between the active site and the HRDC domain.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
1RTV
 
 | | RmlC (dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase) crystal structure from Pseudomonas aeruginosa, apo structure | | Descriptor: | S,R MESO-TARTARIC ACID, dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Dong, C.J, Naismith, J.H. | | Deposit date: | 2003-12-10 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RmlC (dTDP-6-deoxy-D-xylo-4-hexulose 3,5-epimerase) crystal structure from Pseudomonas aeruginosa, apo structure
TO BE PUBLISHED
|
|
2HDJ
 
 | | Crystal structure of human type 3 3alpha-hydroxysteroid dehydrogenase in complex with NADP(H) | | Descriptor: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C2, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Faucher, F, Pereira de Jesus-Tran, K, Cantin, L, Luu-the, V, Labrie, F, Breton, R. | | Deposit date: | 2006-06-20 | | Release date: | 2006-12-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structures of Mouse 17alpha-Hydroxysteroid Dehydrogenase (Apoenzyme and Enzyme-NADP(H) Binary Complex): Identification of Molecular Determinants Responsible for the Unique 17alpha-reductive Activity of this Enzyme.
J.Mol.Biol., 364, 2006
|
|
2HE3
 
 | | Crystal structure of the selenocysteine to cysteine mutant of human glutathionine peroxidase 2 (GPX2) | | Descriptor: | CHLORIDE ION, Glutathione peroxidase 2 | | Authors: | Johansson, C, Kavanagh, K.L, Rojkova, A, Gileadi, O, von Delft, F, Arrowsmith, C, Weigelt, J, Sundstrom, M, Edwards, A, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-06-21 | | Release date: | 2006-07-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the selenocysteine to cysteine mutant of human glutathionine peroxidase 2 (GPX2)
To be Published
|
|
1NRK
 
 | |
1R3K
 
 | | potassium channel KcsA-Fab complex in low concentration of Tl+ | | Descriptor: | Antibody Fab fragment heavy chain, Antibody Fab fragment light chain, DIACYL GLYCEROL, ... | | Authors: | Zhou, Y, MacKinnon, R. | | Deposit date: | 2003-10-02 | | Release date: | 2003-11-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The occupancy of ions in the K+ selectivity filter: Charge balance and coupling of ion binding to a protein conformational change underlie high conduction rates
J.Mol.Biol., 333, 2003
|
|
2H4N
 
 | | H94N CARBONIC ANHYDRASE II COMPLEXED WITH ACETAZOLAMIDE | | Descriptor: | 5-ACETAMIDO-1,3,4-THIADIAZOLE-2-SULFONAMIDE, CARBONIC ANHYDRASE II, ZINC ION | | Authors: | Lesburg, C.A, Christianson, D.W. | | Deposit date: | 1997-05-29 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Histidine --> carboxamide ligand substitutions in the zinc binding site of carbonic anhydrase II alter metal coordination geometry but retain catalytic activity.
Biochemistry, 36, 1997
|
|
1NUS
 
 | | CRYSTAL STRUCTURE OF HUMAN CYTOSOLIC NMN/NaMN ADENYLYLTRANSFERASE COMPLEXED WITH ATP ANALOG AND NMN | | Descriptor: | BETA-NICOTINAMIDE RIBOSE MONOPHOSPHATE, DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, FKSG76, ... | | Authors: | Zhang, X, Kurnasov, O.V, Karthikeyan, S, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2003-02-01 | | Release date: | 2003-06-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | STRUCTURAL CHARACTERIZATION OF A HUMAN CYTOSOLIC NMN/NaMN
ADENYLYLTRANSFERASE AND IMPLICATION IN HUMAN NAD BIOSYNTHESIS
J.Biol.Chem., 278, 2003
|
|
1RA0
 
 | | Bacterial cytosine deaminase D314G mutant bound to 5-fluoro-4-(S)-hydroxy-3,4-dihydropyrimidine. | | Descriptor: | (4S)-5-FLUORO-4-HYDROXY-3,4-DIHYDROPYRIMIDIN-2(1H)-ONE, Cytosine deaminase, FE (III) ION | | Authors: | Mahan, S.D, Ireton, G.C, Stoddard, B.L, Black, M.E. | | Deposit date: | 2003-10-31 | | Release date: | 2004-10-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Random mutagenesis and selection of Escherichia coli cytosine deaminase for cancer gene therapy.
Protein Eng.Des.Sel., 17, 2004
|
|
2HFH
 
 | |
1NUL
 
 | | XPRTASE FROM E. COLI | | Descriptor: | MAGNESIUM ION, SULFATE ION, XANTHINE-GUANINE PHOSPHORIBOSYLTRANSFERASE | | Authors: | Vos, S, De Jersey, J, Martin, J.L. | | Deposit date: | 1996-10-15 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Escherichia coli xanthine phosphoribosyltransferase.
Biochemistry, 36, 1997
|
|
2GLT
 
 | | STRUCTURE OF ESCHERICHIA COLI GLUTATHIONE SYNTHETASE AT PH 6.0. | | Descriptor: | GLUTATHIONE BIOSYNTHETIC LIGASE | | Authors: | Matsuda, K, Yamaguchi, H, Kato, H, Nishioka, T, Katsube, Y, Oda, J. | | Deposit date: | 1995-05-16 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of glutathione synthetase at optimal pH: domain architecture and structural similarity with other proteins.
Protein Eng., 9, 1996
|
|
2HCO
 
 | |
2GTU
 
 | | LIGAND-FREE HUMAN GLUTATHIONE S-TRANSFERASE M2-2 (E.C.2.5.1.18), MONOCLINIC CRYSTAL FORM | | Descriptor: | GLUTATHIONE S-TRANSFERASE | | Authors: | Patskovska, L.N, Fedorov, A.A, Patskovsky, Y.V, Almo, S.C, Listowsky, I. | | Deposit date: | 1998-05-26 | | Release date: | 1999-03-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The enhanced affinity for thiolate anion and activation of enzyme-bound glutathione is governed by an arginine residue of human Mu class glutathione S-transferases.
J.Biol.Chem., 275, 2000
|
|
2GSQ
 
 | |
