1LQA
 
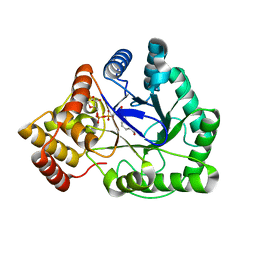 | | TAS PROTEIN FROM ESCHERICHIA COLI IN COMPLEX WITH NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Tas protein | | Authors: | Obmolova, G, Teplyakov, A, Khil, P.P, Howard, A.J, Camerini-Otero, R.D, Gilliland, G.L, Structure 2 Function Project (S2F) | | Deposit date: | 2002-05-09 | | Release date: | 2003-07-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Crystal structure of the Escherichia coli Tas protein, an NADP(H)-dependent aldo-keto reductase
PROTEINS: STRUCT.,FUNCT.,GENET., 53, 2003
|
|
1LH0
 
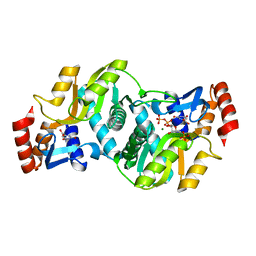 | | Crystal Structure of Salmonella typhimurium OMP Synthase in Complex with MGPRPP and Orotate | | Descriptor: | 1-O-pyrophosphono-5-O-phosphono-alpha-D-ribofuranose, MAGNESIUM ION, OMP synthase, ... | | Authors: | Fedorov, A.A, Panneerselvam, K, Shi, W, Grubmeyer, C, Almo, S.C. | | Deposit date: | 2002-04-16 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Salmonella typhimurium OMP Synthase in a Complete Substrate Complex.
Biochemistry, 51, 2012
|
|
1LQW
 
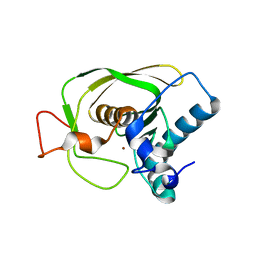 | | Crystal Structure of S.aureus Peptide Deformylase | | Descriptor: | PEPTIDE DEFORMYLASE PDF1, ZINC ION | | Authors: | Mikol, V. | | Deposit date: | 2002-05-14 | | Release date: | 2002-07-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | The crystal structures of four peptide deformylases bound to the antibiotic actinonin reveal two distinct types: a platform for the structure-based design of antibacterial agents.
J.Mol.Biol., 320, 2002
|
|
2JXD
 
 | |
2JUA
 
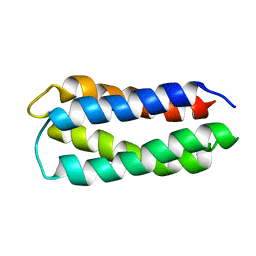 | | Assignment, structure, and dynamics of de novo designed protein S836 | | Descriptor: | de novo protein S836 | | Authors: | Go, A, Kim, S, Baum, J.S, Hecht, M.H. | | Deposit date: | 2007-08-16 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of de novo proteins from a designed superfamily of 4-helix bundles.
Protein Sci., 17, 2008
|
|
1V76
 
 | |
2JYU
 
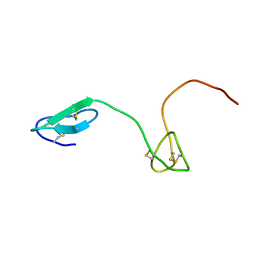 | | Human Granulin C, isomer 2 | | Descriptor: | Granulin-5 | | Authors: | Tolkatchev, D, Wang, P, Chen, Z, Xu, P, Ni, F. | | Deposit date: | 2007-12-19 | | Release date: | 2008-04-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Structure dissection of human progranulin identifies well-folded granulin/epithelin modules with unique functional activities.
Protein Sci., 17, 2008
|
|
2JZF
 
 | | NMR Conformer closest to the mean coordinates of the domain 513-651 of the SARS-CoV nonstructural protein nsp3 | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Chatterjee, A, Johnson, M.A, Serrano, P, Pedrini, B, Joseph, J, Saikatendu, K, Neuman, B, Stevens, R.C, Wilson, I.A, Buchmeier, M.J, Kuhn, P, Wuthrich, K, Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structure shows that the severe acute respiratory syndrome coronavirus-unique domain contains a macrodomain fold.
J.Virol., 83, 2009
|
|
1LKD
 
 | | CRYSTAL STRUCTURE OF 2,3-DIHYDROXYBIPHENYL 1,2-DIOXYGENASE (DHBD) COMPLEXED WITH 2',6'-DICL DIHYDROXYBIPHENYL (DHB) | | Descriptor: | 2',6'-DICHLORO-BIPHENYL-2,6-DIOL, BIPHENYL-2,3-DIOL 1,2-DIOXYGENASE, FE (II) ION, ... | | Authors: | Dai, S, Bolin, J.T. | | Deposit date: | 2002-04-24 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Identification and analysis of a bottleneck in PCB biodegradation
Nat.Struct.Biol., 9, 2002
|
|
1LLI
 
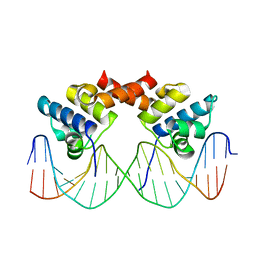 | | THE CRYSTAL STRUCTURE OF A MUTANT PROTEIN WITH ALTERED BUT IMPROVED HYDROPHOBIC CORE PACKING | | Descriptor: | DNA (5'-D(*AP*AP*TP*AP*CP*CP*AP*CP*TP*GP*GP*CP*GP*GP*TP*GP*A P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*TP*CP*AP*CP*CP*GP*CP*CP*AP*GP*TP*GP*G P*TP*AP*T)-3'), PROTEIN (LAMBDA REPRESSOR) | | Authors: | Lim, W.A, Hodel, A, Sauer, R.T, Richards, F.M. | | Deposit date: | 1994-03-25 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The crystal structure of a mutant protein with altered but improved hydrophobic core packing.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
1LLQ
 
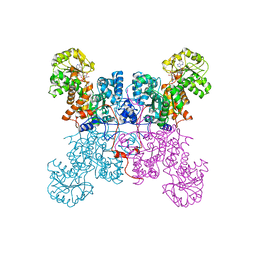 | | Crystal Structure of Malic Enzyme from Ascaris suum Complexed with Nicotinamide Adenine Dinucleotide | | Descriptor: | NAD-dependent malic enzyme, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Coleman, D.E, Jagannatha, G.S, Goldsmith, E.J, Cook, P.F, Harris, B.G. | | Deposit date: | 2002-04-29 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the malic enzyme from Ascaris suum complexed with nicotinamide adenine dinucleotide at 2.3 A resolution.
Biochemistry, 41, 2002
|
|
1URG
 
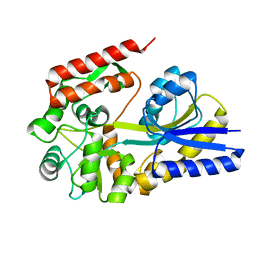 | | X-ray structures from the maltose-maltodextrin binding protein of the thermoacidophilic bacterium Alicyclobacillus acidocaldarius | | Descriptor: | MALTOSE-BINDING PROTEIN, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Schafer, K, Magnusson, U, Scheffel, F, Schiefner, A, Sandgren, M.O.J, Diederichs, K, Welte, W, Hulsmann, A, Schneider, E, Mowbray, S.L. | | Deposit date: | 2003-10-29 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures of the Maltose-Maltodextrin-Binding Protein of the Thermoacidophilic Bacterium Alicyclobacillus Acidocaldarius Provide Insight Into Acid Stability of Proteins.
J.Mol.Biol., 335, 2004
|
|
1KYN
 
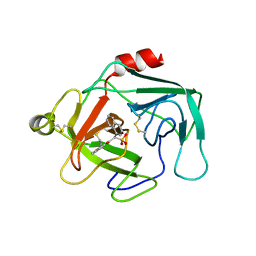 | | Cathepsin-G | | Descriptor: | (2-NAPHTHALEN-2-YL-1-NAPHTHALEN-1-YL-2-OXO-ETHYL)-PHOSPHONIC ACID, cathepsin G | | Authors: | Greco, M.N, Hawkins, M.J, Powell, E.T, Almond Jr, H.R, Corcoran, T.W, De Garavilla, L, Kauffman, J.A, Recacha, R, Chattopadhyay, D, Andrade-Gordon, P, Maryanoff, B.E. | | Deposit date: | 2002-02-05 | | Release date: | 2002-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Nonpeptide inhibitors of cathepsin G: optimization of a novel beta-ketophosphonic acid lead by structure-based drug design.
J.Am.Chem.Soc., 124, 2002
|
|
1KZ9
 
 | | Mutant Enzyme L119F Lumazine Synthase from S.pombe | | Descriptor: | 6,7-Dimethyl-8-ribityllumazine Synthase, PHOSPHATE ION | | Authors: | Gerhardt, S, Haase, I, Steinbacher, S, Kaiser, J.T, Cushman, M, Bacher, A, Huber, R, Fischer, M. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The structural basis of riboflavin binding to Schizosaccharomyces pombe 6,7-dimethyl-8-ribityllumazine synthase.
J.Mol.Biol., 318, 2002
|
|
1L0D
 
 | |
1KZA
 
 | | Complex of MBP-C and Man-a13-Man | | Descriptor: | CALCIUM ION, CHLORIDE ION, MANNOSE-BINDING PROTEIN C, ... | | Authors: | Ng, K.K, Kolatkar, A.R, Park-Snyder, S, Feinberg, H, Clark, D.A, Drickamer, K, Weis, W.I. | | Deposit date: | 2002-02-06 | | Release date: | 2002-07-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Orientation of bound ligands in mannose-binding proteins. Implications for multivalent ligand recognition.
J.Biol.Chem., 277, 2002
|
|
6RRI
 
 | | Human Carbonic Anhydrase II in complex with fluorinated benzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 2,3,5,6-tetrakis(fluoranyl)benzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2019-05-18 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | The Influence of Varying Fluorination Patterns on the Thermodynamics and Kinetics of Benzenesulfonamide Binding to Human Carbonic Anhydrase II.
Biomolecules, 10, 2020
|
|
1L2M
 
 | | Minimized Average Structure of the N-terminal, DNA-binding domain of the replication initiation protein from a geminivirus (Tomato yellow leaf curl virus-Sardinia) | | Descriptor: | Rep protein | | Authors: | Campos-Olivas, R, Louis, J.M, Clerot, D, Gronenborn, B, Gronenborn, A.M. | | Deposit date: | 2002-02-22 | | Release date: | 2002-09-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of a replication initiator unites diverse aspects of nucleic acid metabolism
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2K19
 
 | | NMR solution structure of PisI | | Descriptor: | Putative piscicolin 126 immunity protein | | Authors: | Martin-Visscher, L.A, Sprules, T, Gursky, L.J, Vederas, J.C. | | Deposit date: | 2008-02-25 | | Release date: | 2008-06-17 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of PisI, a group B immunity protein that provides protection against the type IIa bacteriocin piscicolin 126, PisA.
Biochemistry, 47, 2008
|
|
2K03
 
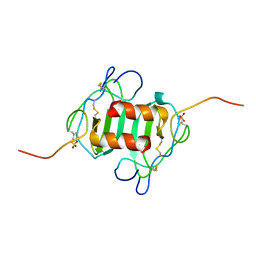 | |
1L0J
 
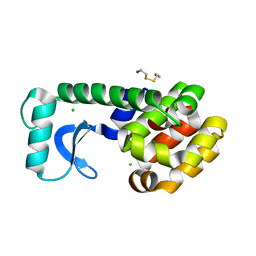 | | METHIONINE CORE MUTANT OF T4 LYSOZYME | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Mooers, B.H, Busam, R.D, Weaver, L.H, Lindstrom, J.D, Quillin, M.L, Matthews, B.W. | | Deposit date: | 2002-02-11 | | Release date: | 2003-06-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Multiple methionine substitutions are tolerated in T4 lysozyme and have coupled effects on folding and stability
BIOPHYS.CHEM., 100, 2003
|
|
2JK1
 
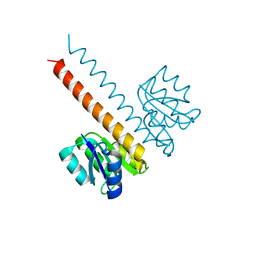 | | Crystal structure of the wild-type HupR receiver domain | | Descriptor: | HYDROGENASE TRANSCRIPTIONAL REGULATORY PROTEIN HUPR1, MAGNESIUM ION | | Authors: | Davies, K.M, Lowe, E.D, Venien-Bryan, C, Johnson, L.N. | | Deposit date: | 2008-05-26 | | Release date: | 2008-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Hupr Receiver Domain Crystal Structure in its Nonphospho and Inhibitory Phospho States.
J.Mol.Biol., 385, 2009
|
|
1L3D
 
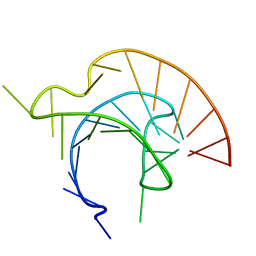 | |
1LIL
 
 | |
2JJ8
 
 | |
