7PZ5
 
 | | Structure of an LPMO at 9.56x10^4 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
7PZ8
 
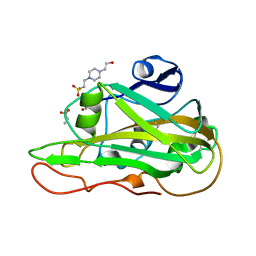 | | Structure of an LPMO at 3.12x10^6 Gy | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACRYLIC ACID, ... | | Authors: | Tandrup, T, Muderspach, S.J, Ipsen, J.O, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
6WBQ
 
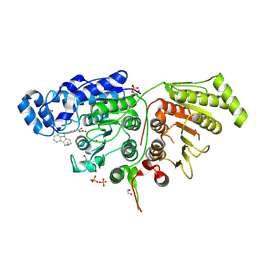 | | Crystal Structure of Danio rerio Histone Deacetylase 10 in Complex with Tubastatin A | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-methyl-3,4-dihydro-1~{H}-pyrido[4,3-b]indol-5-yl)methyl]-~{N}-oxidanyl-benzamide, PHOSPHATE ION, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-03-27 | | Release date: | 2020-07-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for the Selective Inhibition of HDAC10, the Cytosolic Polyamine Deacetylase.
Acs Chem.Biol., 15, 2020
|
|
8UHH
 
 | | anti-Phosphohistidine Fab hSC44.ck.elbow.20.N32F | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Kalagiri, R, Stanfield, R.L, Hunter, T, Wilson, I.A. | | Deposit date: | 2023-10-09 | | Release date: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Using phage display for rational engineering of a higher affinity humanized 3' phosphohistidine-specific antibody.
Biorxiv, 2024
|
|
7YT0
 
 | |
8DWK
 
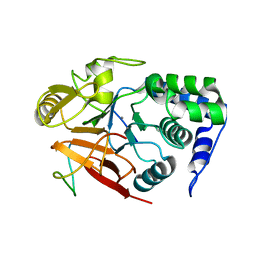 | | Inhibitor-3:PP1 reconstituted complex | | Descriptor: | E3 ubiquitin-protein ligase PPP1R11, MANGANESE (II) ION, Serine/threonine-protein phosphatase PP1-alpha catalytic subunit | | Authors: | Choy, M.S, Srivastava, G, Page, R, Peti, W. | | Deposit date: | 2022-08-01 | | Release date: | 2023-02-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor-3 inhibits Protein Phosphatase 1 via a metal binding dynamic protein-protein interaction.
Nat Commun, 14, 2023
|
|
6VQZ
 
 | | HLA-B*27:05 presenting an HIV-1 6mer peptide | | Descriptor: | 6-mer peptide, ARGININE, Beta-2-microglobulin, ... | | Authors: | Pymm, P, Tenzer, S, Wee, E, Weimershaus, M, Burgevin, A, Kollnberger, S, Gerstoft, J, Josephs, T.M, Ladell, K, Mclaren, J.E, Appay, V, Price, D.A, Fugger, L, Bell, J.I, Hansjorg, S, Van Endert, P, Harkiolaki, M, Iversen, A.K.N. | | Deposit date: | 2020-02-06 | | Release date: | 2021-02-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Epitope length variants balance protective immune responses and viral escape in HIV-1 infection
Cell Rep, 38, 2022
|
|
1EN1
 
 | | STRUCTURE OF THE HIV-1 MINUS STRAND PRIMER BINDING SITE | | Descriptor: | DNA (5'-D(P*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*A)-3') | | Authors: | Johnson, P.E, Turner, R.B, Wu, Z.R, Levin, J.G, Summers, M.F. | | Deposit date: | 2000-03-20 | | Release date: | 2000-04-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A mechanism for plus-strand transfer enhancement by the HIV-1 nucleocapsid protein during reverse transcription
Biochemistry, 39, 2000
|
|
8DRK
 
 | | LRRC8A:C conformation 1 (round) top focus | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, Volume-regulated anion channel subunit LRRC8A,Soluble cytochrome b562, Volume-regulated anion channel subunit LRRC8C | | Authors: | Kern, D.M, Brohawn, S.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-03-08 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (2.95 Å) | | Cite: | Structural basis for assembly and lipid-mediated gating of LRRC8A:C volume-regulated anion channels.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4WKH
 
 | | Crystal structure of human chitotriosidase-1 catalytic domain in complex with chitobiose (1mM) at 1.05 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitotriosidase-1 | | Authors: | Fadel, F, Zhao, Y, Cachau, R, Cousido-Siah, A, Ruiz, F.X, Harlos, K, Howard, E, Mitschler, A, Podjarny, A. | | Deposit date: | 2014-10-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | New insights into the enzymatic mechanism of human chitotriosidase (CHIT1) catalytic domain by atomic resolution X-ray diffraction and hybrid QM/MM.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WKV
 
 | | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ | | Descriptor: | Acyl-homoserine lactone acylase PvdQ, GLYCEROL, trihydroxy(octyl)borate(1-) | | Authors: | Wu, R, Clevenger, K.D, Fast, W, Liu, D. | | Deposit date: | 2014-10-03 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1434 Å) | | Cite: | n-Alkylboronic Acid Inhibitors Reveal Determinants of Ligand Specificity in the Quorum-Quenching and Siderophore Biosynthetic Enzyme PvdQ.
Biochemistry, 53, 2014
|
|
6W0U
 
 | |
6W2I
 
 | | Crystal Structure of Y188G Variant of the Internal UBA Domain of HHR23A | | Descriptor: | GLYCEROL, UV excision repair protein RAD23 homolog A | | Authors: | Bowler, B.E, Zeng, B, Becht, D.C, Rothfuss, M, Sprang, S.R, Mou, T.-C. | | Deposit date: | 2020-03-05 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | High-Accuracy Prediction of Stabilizing Surface Mutations to the Three-Helix Bundle, UBA(1), with EmCAST.
J.Am.Chem.Soc., 145, 2023
|
|
6WC8
 
 | |
8QDB
 
 | | Wdyg1p Ser256DHA (PSF) | | Descriptor: | 1,2-ETHANEDIOL, Yellowish-green 1-like protein | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8CHD
 
 | | NtUGT1 in two conformations | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Glycosyltransferase, ... | | Authors: | Fredslund, F, Teze, D, Adams, P.D, Welner, D.H. | | Deposit date: | 2023-02-07 | | Release date: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | NtUGT1 in two conformations
To Be Published
|
|
1EBS
 
 | |
8VKZ
 
 | | Crystal structure of Glucocorticoid Receptor in complex with an inhibitor | | Descriptor: | (4aR,4bS,5R,6aS,6bS,8R,9aR,10aR,10bR)-8-{4-[(3-aminophenyl)methyl]phenyl}-5-hydroxy-6b-(hydroxyacetyl)-4a,6a-dimethyl-4a,4b,5,6,6a,6b,9a,10,10a,10b,11,12-dodecahydro-2H,8H-naphtho[2',1':4,5]indeno[1,2-d][1,3]dioxol-2-one, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | Judge, R.A, Hobson, A.D. | | Deposit date: | 2024-01-10 | | Release date: | 2024-01-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.133 Å) | | Cite: | Minimising the payload solvent exposed hydrophobic surface area optimises the antibody-drug conjugate properties.
Rsc Med Chem, 15, 2024
|
|
1EBR
 
 | |
9O0Y
 
 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, N-[(2,3-dichlorophenyl)methyl]-3,5-difluoro-N-[(pyridin-3-yl)methyl]benzamide, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2025-04-03 | | Release date: | 2025-05-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
1EBQ
 
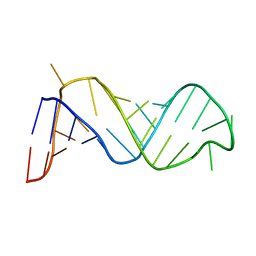 | |
6VR8
 
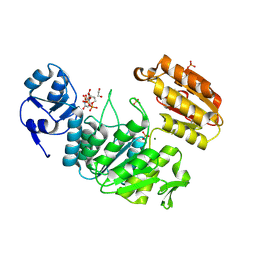 | | Structure of a pseudomurein peptide ligase type E from Methanothermus fervidus | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Mur ligase middle domain protein, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S, Subedi, B.P. | | Deposit date: | 2020-02-06 | | Release date: | 2021-08-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterisation of methanogen pseudomurein cell wall peptide ligases homologous to bacterial MurE/F murein peptide ligases.
Microbiology (Reading, Engl.), 168, 2022
|
|
8QBI
 
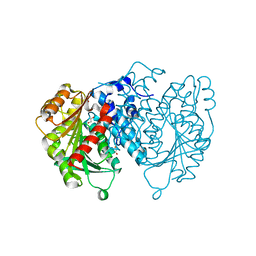 | | AntI in closed state | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Photorhabdus luminescens subsp. laumondii TTO1 complete genome segment 15/17, ... | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-24 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
8CU5
 
 | |
4X12
 
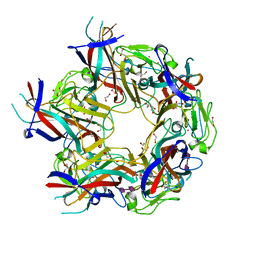 | | JC Polyomavirus genotype 3 VP1 in complex with GD1b oligosaccharide | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein, ... | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-11-24 | | Release date: | 2015-04-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Greater Affinity of JC Polyomavirus Capsid for alpha 2,6-Linked Lactoseries Tetrasaccharide c than for Other Sialylated Glycans Is a Major Determinant of Infectivity.
J.Virol., 89, 2015
|
|
