134D
 
 | |
1ONC
 
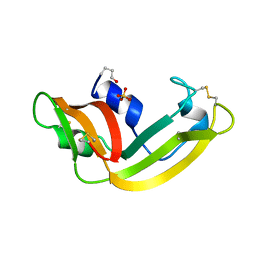 | | THE REFINED 1.7 ANGSTROMS X-RAY CRYSTALLOGRAPHIC STRUCTURE OF P-30, AN AMPHIBIAN RIBONUCLEASE WITH ANTI-TUMOR ACTIVITY | | Descriptor: | P-30 PROTEIN, SULFATE ION | | Authors: | Mosimann, S.C, Ardelt, W, James, M.N.G. | | Deposit date: | 1993-08-30 | | Release date: | 1994-01-31 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined 1.7 A X-ray crystallographic structure of P-30 protein, an amphibian ribonuclease with anti-tumor activity.
J.Mol.Biol., 236, 1994
|
|
1PKP
 
 | |
135D
 
 | |
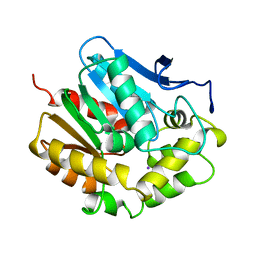
2EDA
 
 | |
2BFH
 
 | | CRYSTAL STRUCTURE OF BASIC FIBROBLAST GROWTH FACTOR AT 1.6 ANGSTROMS RESOLUTION | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Kitagawa, Y, Ago, H, Katsube, Y, Fujishima, A, Matsuura, Y. | | Deposit date: | 1993-08-31 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of basic fibroblast growth factor at 1.6 A resolution.
J.Biochem.(Tokyo), 110, 1991
|
|
1RPB
 
 | | SOLUTION STRUCTURE OF RP 71955, A NEW 21 AMINO ACID TRICYCLIC PEPTIDE ACTIVE AGAINST HIV-1 VIRUS | | Descriptor: | Tricyclic peptide RP 71955 | | Authors: | Frechet, D, Guitton, J.D, Herman, F, Faucher, D, Helynck, G, Monegier Du Sorbier, B, Ridoux, J.P, James-Surcouf, E, Vuilhorgne, M. | | Deposit date: | 1993-08-31 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RP 71955, a new 21 amino acid tricyclic peptide active against HIV-1 virus.
Biochemistry, 33, 1994
|
|
1RPC
 
 | | SOLUTION STRUCTURE OF RP 71955, A NEW 21 AMINO ACID TRICYCLIC PEPTIDE ACTIVE AGAINST HIV-1 VIRUS | | Descriptor: | Tricyclic peptide RP 71955 | | Authors: | Frechet, D, Guitton, J.D, Herman, F, Faucher, D, Helynck, G, Monegier Du Sorbier, B, Ridoux, J.P, James-Surcouf, E, Vuilhorgne, M. | | Deposit date: | 1993-08-31 | | Release date: | 1994-01-31 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure of RP 71955, a new 21 amino acid tricyclic peptide active against HIV-1 virus.
Biochemistry, 33, 1994
|
|
1BCD
 
 | |
1GDR
 
 | |
2EDC
 
 | |
138L
 
 | |
139L
 
 | |
8TLN
 
 | |
1EGL
 
 | |
2AAI
 
 | | Crystallographic refinement of ricin to 2.5 Angstroms | | Descriptor: | RICIN (A CHAIN), RICIN (B CHAIN), alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rutenber, E, Katzin, B.J, Montfort, W, Villafranca, J.E, Ernst, S.R, Collins, E.J, Mlsna, D, Monzingo, A.F, Ready, M.P, Robertus, J.D. | | Deposit date: | 1993-09-07 | | Release date: | 1994-01-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic refinement of ricin to 2.5 A.
Proteins, 10, 1991
|
|
2RSL
 
 | |
2DHD
 
 | |
2DHC
 
 | |
1GSB
 
 | |
1GSC
 
 | |
1OMB
 
 | |
1OMA
 
 | |
1NAR
 
 | |
1MAP
 
 | |
