8VH2
 
 | | CH235.12 Fab bound to the HIV-1 CH505.M5 SOSIP | | Descriptor: | CH235.12 Fab Heavy Chain, CH235.12 Fab Light Chain, CH505.M5.G458Y SOSIP gp120, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-02 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies
To Be Published
|
|
8VGW
 
 | | VRC01 Fab bound to the HIV-1 CH848 DE3 SOSIP | | Descriptor: | CH848 DE3 SOSIP gp120, CH848 DE3 SOSIP gp41, VRC01 Fab Heavy Chain, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-29 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies
To Be Published
|
|
1A8I
 
 | | SPIROHYDANTOIN INHIBITOR OF GLYCOGEN PHOSPHORYLASE | | Descriptor: | BETA-D-GLUCOPYRANOSE SPIROHYDANTOIN, GLYCOGEN PHOSPHORYLASE B | | Authors: | Gregoriou, M, Noble, M.E.M, Watson, K.A, Garman, E.F, Krulle, T.M, De La Fuente, C, Fleet, G.W.J, Oikonomakos, N.G, Johnson, L.N. | | Deposit date: | 1998-03-25 | | Release date: | 1998-07-01 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The structure of a glycogen phosphorylase glucopyranose spirohydantoin complex at 1.8 A resolution and 100 K: the role of the water structure and its contribution to binding.
Protein Sci., 7, 1998
|
|
2LJE
 
 | |
8VH1
 
 | | CH235.12 Fab bound to the HIV-1 CH505.M5 SOSIP | | Descriptor: | CH235.I60 Fab Heavy Chain, CH235.I60 Fab Light Chain, CH505.M5 SOSIP gp120, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-30 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies
To Be Published
|
|
8VGV
 
 | | DH270.6 Fab bound to the HIV-1 CH848 DE3 SOSIP | | Descriptor: | CH848 DE3 SOSIP gp120, CH848 DE3 SOSIP gp41, DH270.6 Antibody Fab Heavy Chain, ... | | Authors: | Henderson, R, Acharya, P. | | Deposit date: | 2023-12-29 | | Release date: | 2024-10-02 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Engineering immunogens that select for specific mutations in HIV broadly neutralizing antibodies
To Be Published
|
|
2R0K
 
 | | Protease domain of HGFA with inhibitor Fab58 | | Descriptor: | Hepatocyte growth factor activator, antibody heavy chain of Fab58, Fab portion only, ... | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2007-08-20 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Structural insight into distinct mechanisms of protease inhibition by antibodies.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4WQA
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - tetrathionate co-crystallization | | Descriptor: | 1,2-ETHANEDIOL, HEME C, IODIDE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
8BZZ
 
 | |
8A3L
 
 | | Structural insights into the binding of bS1 to the ribosome | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S1, ... | | Authors: | D'Urso, G, Chat, S, Gillet, R, Giudice, E. | | Deposit date: | 2022-06-08 | | Release date: | 2023-05-10 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural insights into the binding of bS1 to the ribosome.
Nucleic Acids Res., 51, 2023
|
|
1EYM
 
 | | FK506 BINDING PROTEIN MUTANT, HOMODIMERIC COMPLEX | | Descriptor: | FK506 BINDING PROTEIN | | Authors: | Rollins, C.T, Rivera, V.M, Woolfson, D.N, Keenan, T, Hatada, M, Adams, S.E, Andrade, L.J, Yaeger, D, van Schravendijk, M.R, Holt, D.A, Gilman, M, Clackson, T. | | Deposit date: | 2000-05-07 | | Release date: | 2000-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A ligand-reversible dimerization system for controlling protein-protein interactions.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
4WGG
 
 | | STRUCTURE OF THE TERNARY COMPLEX OF A ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE IN COMPLEX WITH NADP AND CONIFERYL ALDEHYDE | | Descriptor: | (2E)-3-(4-hydroxy-3-methoxyphenyl)prop-2-enal, Double Bond Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Collery, J, Langlois d'Estaintot, B, Buratto, J, Granier, T, Gallois, B, Willis, M.A, Sang, Y, Flores-Sanchez, I.J, Gang, D.R. | | Deposit date: | 2014-09-18 | | Release date: | 2015-09-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | STRUCTURE OF ZINGIBER OFFICINALE DOUBLE BOND REDUCTASE
to be published
|
|
4WRN
 
 | | Crystal structure of the polymerization region of human uromodulin/Tamm-Horsfall protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Uromodulin, ZINC ION, ... | | Authors: | Bokhove, M, De Sanctis, D, Jovine, L. | | Deposit date: | 2014-10-24 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A structured interdomain linker directs self-polymerization of human uromodulin.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4WRV
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with uracil, Form III | | Descriptor: | CHLORIDE ION, URACIL, Uracil-DNA glycosylase | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WRZ
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-fluorouracil (AB), Form I | | Descriptor: | 5-FLUOROURACIL, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WS2
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 6-aminouracil, Form I | | Descriptor: | 6-aminopyrimidine-2,4(3H,5H)-dione, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WS7
 
 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with 5-chlorouracil, Form II | | Descriptor: | 1,2-ETHANEDIOL, 5-chloropyrimidine-2,4(1H,3H)-dione, CHLORIDE ION, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-25 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1B6Z
 
 | | 6-PYRUVOYL TETRAHYDROPTERIN SYNTHASE | | Descriptor: | 6-pyruvoyl tetrahydropterin synthase, ZINC ION | | Authors: | Ploom, T, Thoeny, B, Yim, J, Lee, S, Nar, H, Leimbacher, W, Huber, R, Richardson, J, Auerbach, G. | | Deposit date: | 1999-01-18 | | Release date: | 2000-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic and kinetic investigations on the mechanism of 6-pyruvoyl tetrahydropterin synthase.
J.Mol.Biol., 286, 1999
|
|
2LSY
 
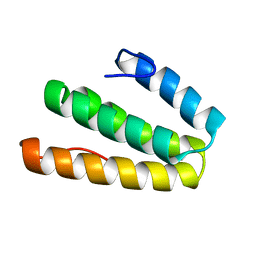 | |
8EVD
 
 | |
1AT1
 
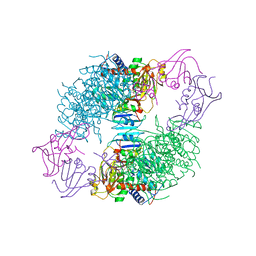 | |
1BCR
 
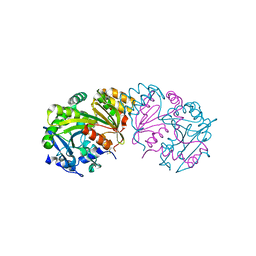 | | COMPLEX OF THE WHEAT SERINE CARBOXYPEPTIDASE, CPDW-II, WITH THE MICROBIAL PEPTIDE ALDEHYDE INHIBITOR, ANTIPAIN, AND ARGININE AT ROOM TEMPERATURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIPAIN, ... | | Authors: | Bullock, T.L, Remington, S.J. | | Deposit date: | 1995-11-03 | | Release date: | 1996-03-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Peptide aldehyde complexes with wheat serine carboxypeptidase II: implications for the catalytic mechanism and substrate specificity.
J.Mol.Biol., 255, 1996
|
|
4WPL
 
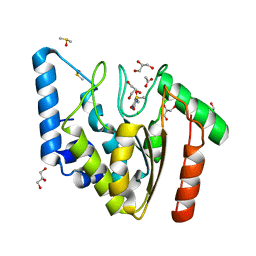 | | Crystal structure of Mycobacterium tuberculosis uracil-DNA glycosylase in complex with uracil, Form I | | Descriptor: | ACETATE ION, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Arif, S.M, Geethanandan, K, Mishra, P, Surolia, A, Varshney, U, Vijayan, M. | | Deposit date: | 2014-10-20 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structural plasticity in Mycobacterium tuberculosis uracil-DNA glycosylase (MtUng) and its functional implications.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4WQ8
 
 | | Thiosulfate dehydrogenase (TsdA) from Allochromatium vinosum - tetrathionate soak | | Descriptor: | HEME C, IODIDE ION, SULFATE ION, ... | | Authors: | Brito, J.A, Denkmann, K, Pereira, I.A.C, Dahl, C, Archer, M. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-18 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.3989 Å) | | Cite: | Thiosulfate Dehydrogenase (TsdA) from Allochromatium vinosum: STRUCTURAL AND FUNCTIONAL INSIGHTS INTO THIOSULFATE OXIDATION.
J.Biol.Chem., 290, 2015
|
|
1IKQ
 
 | | Pseudomonas Aeruginosa Exotoxin A, wild type | | Descriptor: | CHLORIDE ION, EXOTOXIN A, SODIUM ION | | Authors: | McKay, D.B, Wedekind, J.E, Trame, C.B. | | Deposit date: | 2001-05-04 | | Release date: | 2001-12-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Refined Crystallographic Structure of Pseudomonas aeruginosa Exotoxin A
and its Implications for the Molecular Mechanism of Toxicity
J.Mol.Biol., 314, 2001
|
|
