5EHW
 
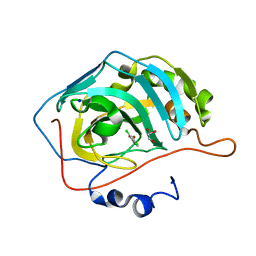 | | human carbonic anhydrase II in complex with ligand | | Descriptor: | (~{E})-3-(2,4-dichlorophenyl)prop-2-enoic acid, Carbonic anhydrase 2, FORMIC ACID, ... | | Authors: | Ren, B. | | Deposit date: | 2015-10-29 | | Release date: | 2016-03-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.392 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance, and X-ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
5WSC
 
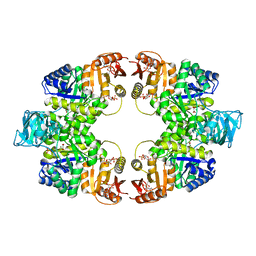 | | Crystal of pyruvate kinase (PYK) from Mycobacterium tuberculosis in complex with Oxalate, soaked with allosteric activators AMP and Glucose 6-Phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Zhong, W, Cai, Q, El Sahili, A, Lescar, J, Dedon, P.C. | | Deposit date: | 2016-12-06 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Allosteric pyruvate kinase-based "logic gate" synergistically senses energy and sugar levels in Mycobacterium tuberculosis.
Nat Commun, 8, 2017
|
|
5FHC
 
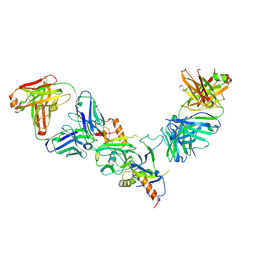 | |
8PGM
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3062 | | Descriptor: | 3-[4-[(azanyl-methyl-oxidanyl-$l^{4}-sulfanyl)methyl]-3-fluoranyl-phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, DIMETHYL SULFOXIDE, ... | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.27 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3062
To Be Published
|
|
5FI3
 
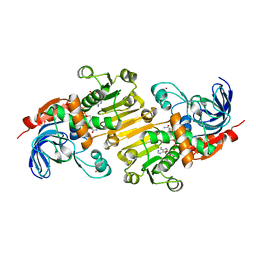 | | HETEROYOHIMBINE SYNTHASE THAS1 FROM CATHARANTHUS ROSEUS - COMPLEX WITH NADP+ | | Descriptor: | 1,2-ETHANEDIOL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Tetrahydroalstonine synthase, ... | | Authors: | Stavrinides, A, Tatsis, E.C, Caputi, L, Foureau, E, Stevenson, C.E.M, Lawson, D.M, Courdavault, V, O'Connor, S.E. | | Deposit date: | 2015-12-22 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural investigation of heteroyohimbine alkaloid synthesis reveals active site elements that control stereoselectivity.
Nat Commun, 7, 2016
|
|
8PGZ
 
 | | Crystal structure of the metallo-beta-lactamase VIM1 with 3528 | | Descriptor: | 7-[(1~{S})-1-[2-(aminomethyl)-6-oxidanylidene-5-oxa-7-azaspiro[3.4]octan-7-yl]ethyl]-3-(6-oxidanylidene-1~{H}-pyridazin-4-yl)-1~{H}-indole-2-carboxylic acid, Beta-lactamase VIM-1, ZINC ION | | Authors: | Calvopina, K, Brem, J, Farley, A.J.M, Allen, M.D, Schofield, C.J. | | Deposit date: | 2023-06-18 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of the metallo-beta-lactamase VIM1 with 3528
To Be Published
|
|
7E46
 
 | |
5FJ2
 
 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with 7-((4-CHLORO-3-((METHYLAMINO)METHYL) PHENOXY)METHYL)QUINOLIN-2-AMINE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[[4-chloranyl-3-(methylaminomethyl)phenoxy]methyl]quinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-10-06 | | Release date: | 2015-10-28 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Phenyl Ether- and Aniline-Containing 2-Aminoquinolines as Potent and Selective Inhibitors of Neuronal Nitric Oxide Synthase.
J.Med.Chem., 58, 2015
|
|
8PJT
 
 | |
7EKZ
 
 | | Structural and functional insights into Hydra Actinoporin-Like Toxin 1 (HALT-1) | | Descriptor: | FORMIC ACID, GLYCEROL, HALT-1 | | Authors: | Ker, D.S, Sha, X.H, Jonet, M.A, Hwang, J.S, Ng, C.L. | | Deposit date: | 2021-04-07 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural and functional analysis of Hydra Actinoporin-Like Toxin 1 (HALT-1).
Sci Rep, 11, 2021
|
|
6WYC
 
 | |
5LVH
 
 | |
7E1C
 
 | | Structure of MreB3 from Spiroplasma eriocheiris | | Descriptor: | ACETATE ION, CALCIUM ION, Cell shape-determining protein MreB | | Authors: | Takahashi, D, Miyata, M, Imada, K. | | Deposit date: | 2021-02-01 | | Release date: | 2022-02-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | ATP-dependent polymerization dynamics of bacterial actin proteins involved in Spiroplasma swimming.
Open Biology, 12, 2022
|
|
6WYQ
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 1 (CD1) K330L mutant complexed with 4-iodo-SAHA | | Descriptor: | Histone deacetylase 6, N~1~-hydroxy-N~8~-(4-iodophenyl)octanediamide, POTASSIUM ION, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.90001464 Å) | | Cite: | Binding of inhibitors to active-site mutants of CD1, the enigmatic catalytic domain of histone deacetylase 6.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5M19
 
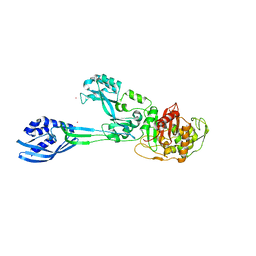 | | Crystal structure of PBP2a from MRSA in the presence of Oxacillin ligand | | Descriptor: | CADMIUM ION, Penicillin-binding protein 2, beta-muramic acid | | Authors: | Molina, R, Batuecas, M.T, Hermoso, J.A. | | Deposit date: | 2016-10-07 | | Release date: | 2017-02-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformational Dynamics in Penicillin-Binding Protein 2a of Methicillin-Resistant Staphylococcus aureus, Allosteric Communication Network and Enablement of Catalysis.
J. Am. Chem. Soc., 139, 2017
|
|
6WZ0
 
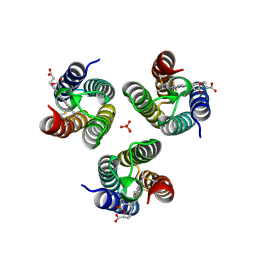 | |
8PJS
 
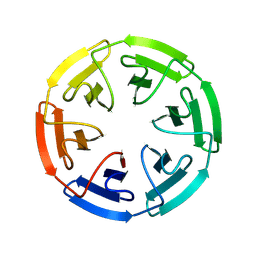 | |
5FJI
 
 | | Three-dimensional structures of two heavily N-glycosylated Aspergillus sp. Family GH3 beta-D-glucosidases | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, BETA-GLUCOSIDASE, ... | | Authors: | Agirre, J, Ariza, A, Offen, W.A, Turkenburg, J.P, Roberts, S.M, McNicholas, S, Harris, P.V, McBrayer, B, Dohnalek, J, Cowtan, K.D, Davies, G.J, Wilson, K.S. | | Deposit date: | 2015-10-09 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Three-Dimensional Structures of Two Heavily N-Glycosylated Aspergillus Sp. Family Gh3 Beta-D-Glucosidases
Acta Crystallogr.,Sect.D, 72, 2016
|
|
6O7O
 
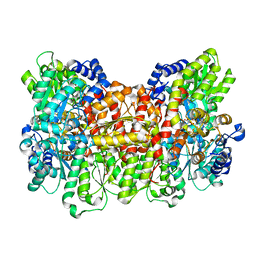 | |
8PMV
 
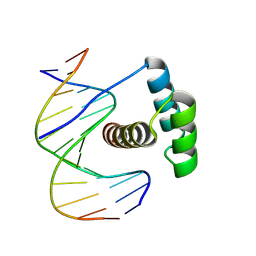 | |
5FLO
 
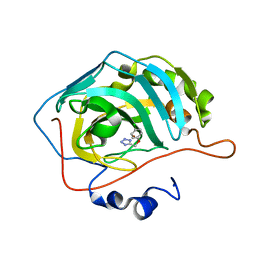 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | 5-[(4-chloranylphenoxy)methyl]-1H-1,2,3,4-tetrazole, CARBONIC ANHYDRASE 2, ZINC ION | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
8PWE
 
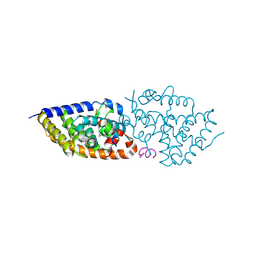 | | Crystal structure of VDR complex with Novel Des-C-Ring and Aromatic-D-Ring analog 3a | | Descriptor: | (1~{R},3~{S},5~{Z})-4-methylidene-5-[(~{E})-3-[3-(6-methyl-6-oxidanyl-hept-3-ynyl)phenyl]pent-2-enylidene]cyclohexane-1,3-diol, ACETATE ION, Nuclear receptor coactivator 2, ... | | Authors: | Rochel, N. | | Deposit date: | 2023-07-20 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Des-C-Ring and Aromatic-D-Ring analogs Acting as Potent Agonists of the Vitamin D Receptor (VDR)
To Be Published
|
|
5FLR
 
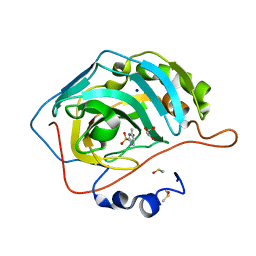 | | Native state mass spectrometry, surface plasmon resonance and X-ray crystallography correlate strongly as a fragment screening combination | | Descriptor: | 1.7.6 4-methylpyrimidine-2-sulfonamide, CARBONIC ANHYDRASE 2, DIMETHYL SULFOXIDE, ... | | Authors: | Woods, L.A, Dolezal, O, Ren, B, Ryan, J.H, Peat, T.S, Poulsen, S.A. | | Deposit date: | 2015-10-28 | | Release date: | 2016-03-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Native State Mass Spectrometry, Surface Plasmon Resonance and X-Ray Crystallography Correlate Strongly as a Fragment Screening Combination.
J.Med.Chem., 59, 2016
|
|
5WUC
 
 | | Structural basis for conductance through TRIC cation channels | | Descriptor: | SODIUM ION, Uncharacterized protein | | Authors: | Su, M, Gao, F, Mao, Y, Li, D.L, Guo, Y.Z, Wang, X.H, Bruni, R, Kloss, B, Hendrickson, W.A, Chen, Y.H, New York Consortium on Membrane Protein Structure (NYCOMPS) | | Deposit date: | 2016-12-17 | | Release date: | 2017-07-12 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis for conductance through TRIC cation channels.
Nat Commun, 8, 2017
|
|
5FK2
 
 | |
