1YTG
 
 | | SIV PROTEASE CRYSTALLIZED WITH PEPTIDE PRODUCT | | Descriptor: | HIV PROTEASE, PEPTIDE PRODUCT | | Authors: | Rose, R.B, Craik, C.S, Douglas, N.L, Stroud, R.M. | | Deposit date: | 1996-08-01 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Three-dimensional structures of HIV-1 and SIV protease product complexes.
Biochemistry, 35, 1996
|
|
1YBV
 
 | | STRUCTURE OF TRIHYDROXYNAPHTHALENE REDUCTASE IN COMPLEX WITH NADPH AND AN ACTIVE SITE INHIBITOR | | Descriptor: | 5-METHYL-1,2,4-TRIAZOLO[3,4-B]BENZOTHIAZOLE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TRIHYDROXYNAPHTHALENE REDUCTASE | | Authors: | Andersson, A, Schneider, G, Lindqvist, Y. | | Deposit date: | 1996-09-23 | | Release date: | 1997-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the ternary complex of 1,3,8-trihydroxynaphthalene reductase from Magnaporthe grisea with NADPH and an active-site inhibitor.
Structure, 4, 1996
|
|
2BEN
 
 | |
2C0U
 
 | | Crystal Structure of a Covalent Complex of Nitroalkane Oxidase Trapped During Substrate Turnover | | Descriptor: | (2S)-2-NITROBUTANE, FLAVIN-ADENINE DINUCLEOTIDE, NITROALKANE OXIDASE | | Authors: | Nagpal, A, Valley, M.P, Fitzpatrick, P.F, Orville, A.M. | | Deposit date: | 2005-09-07 | | Release date: | 2006-02-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structures of Nitroalkane Oxidase: Insights Into the Reaction Mechanism from a Covalent Complex of the Flavoenzyme Trapped During Turnover.
Biochemistry, 45, 2006
|
|
1YMU
 
 | |
2BG1
 
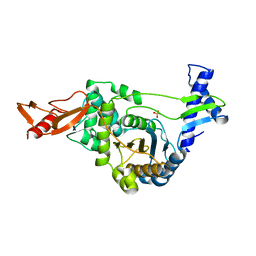 | | Active site restructuring regulates ligand recognition in classA Penicillin-binding proteins (PBPs) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SULFATE ION | | Authors: | Macheboeuf, P, Di Guilmi, A.M, Job, V, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2004-12-16 | | Release date: | 2005-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Active Site Restructuring Regulates Ligand Recognition in Class a Penicillin-Binding Proteins
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BLI
 
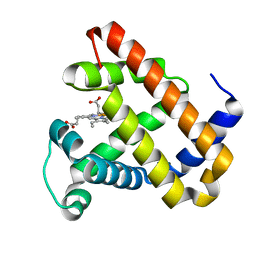 | | L29W Mb deoxy | | Descriptor: | MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
1XGP
 
 | |
2BW9
 
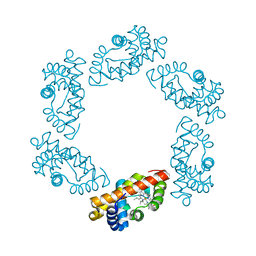 | | Laue Structure of L29W MbCO | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Schmidt, M, Nienhaus, K, Pahl, R, Krasselt, A, Anderson, S, Parak, F, Nienhaus, G.U, Srajer, V. | | Deposit date: | 2005-07-13 | | Release date: | 2005-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Ligand Migration Pathway and Protein Dynamics in Myoglobin: A Time-Resolved Crystallographic Study on L29W Mbco.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BXV
 
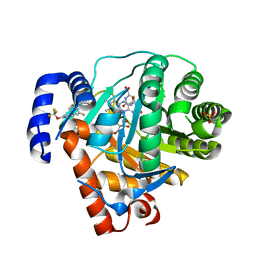 | | Dual binding mode of a novel series of DHODH inhibitors | | Descriptor: | 2-({[3-FLUORO-3'-(TRIFLUOROMETHOXY)BIPHENYL-4-YL]AMINO}CARBONYL)CYCLOPENT-1-ENE-1-CARBOXYLIC ACID, ACETATE ION, DIHYDROOROTATE DEHYDROGENASE, ... | | Authors: | Baumgartner, R, Walloschek, M, Karlik, M, Gotschlich, A, Tasler, S, Mies, J, Leban, J. | | Deposit date: | 2005-07-27 | | Release date: | 2006-08-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Dual binding mode of a novel series of DHODH inhibitors.
J. Med. Chem., 49, 2006
|
|
1U2Z
 
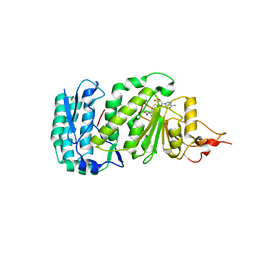 | | Crystal structure of histone K79 methyltransferase Dot1p from yeast | | Descriptor: | Histone-lysine N-methyltransferase, H3 lysine-79 specific, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Sawada, K, Yang, Z, Horton, J.R, Collins, R.E, Zhang, X, Cheng, X. | | Deposit date: | 2004-07-20 | | Release date: | 2004-09-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the conserved core of the yeast Dot1p, a nucleosomal histone H3 lysine 79 methyltransferase
J.Biol.Chem., 279, 2004
|
|
2BZW
 
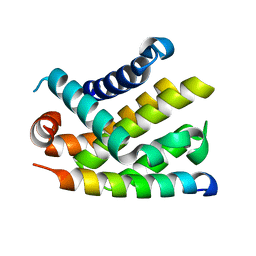 | | The crystal structure of BCL-XL in complex with full-length BAD | | Descriptor: | APOPTOSIS REGULATOR BCL-X, BCL2-ANTAGONIST OF CELL DEATH | | Authors: | Lee, K.-H, Han, W.-D, Kim, K.-J, Oh, B.-H. | | Deposit date: | 2005-08-24 | | Release date: | 2007-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Biochemical Bases for the Inhibition of Autophagy and Apoptosis by Viral Bcl-2 of Murine Gamma-Herpesvirus 68.
Plos Pathog., 4, 2008
|
|
2BU1
 
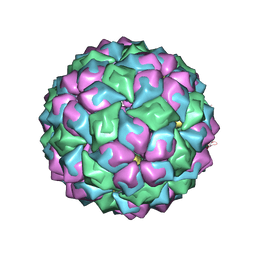 | | MS2-RNA HAIRPIN (5BRU -5) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*GP*AP*GP*GP*AP*UP *5BU*AP*CP*CP*CP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Grahn, E, Moss, T, Helgstrand, C, Fridborg, K, Sundaram, M, Tars, K, Lago, H, Stonehouse, N.J, Davis, D.R, Stockley, P.G, Liljas, L. | | Deposit date: | 2005-06-08 | | Release date: | 2005-08-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of pyrimidine specificity in the MS2 RNA hairpin-coat-protein complex.
Rna, 7, 2001
|
|
1XJD
 
 | |
2C56
 
 | | A comparative study of uracil DNA glycosylases from human and herpes simplex virus type 1 | | Descriptor: | URACIL DNA GLYCOSYLASE, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Krusong, K, Carpenter, E.P, Bellamy, S.R.W, Savva, R, Baldwin, G.S. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Comparative Study of Uracil-DNA Glycosylases from Human and Herpes Simplex Virus Type 1.
J.Biol.Chem., 281, 2006
|
|
2C5F
 
 | | Torpedo californica acetylcholinesterase in complex with a non hydrolysable substrate analogue, 4-oxo-N,N,N-trimethylpentanaminium | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4,4-DIHYDROXY-N,N,N-TRIMETHYLPENTAN-1-AMINIUM, ACETYLCHOLINESTERASE, ... | | Authors: | Colletier, J.P, Fournier, D, Greenblatt, H.M, Sussman, J.L, Zaccai, G, Silman, I, Weik, M. | | Deposit date: | 2005-10-27 | | Release date: | 2006-06-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights Into Substrate Traffic and Inhibition in Acetylcholinesterase.
Embo J., 25, 2006
|
|
2C5D
 
 | | Structure of a minimal Gas6-Axl complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GROWTH-ARREST-SPECIFIC PROTEIN 6 PRECURSOR, ... | | Authors: | Sasaki, T, Knyazev, P.G, Clout, N.J, Cheburkin, Y, Goehring, W, Ullrich, A, Timpl, R, Hohenester, E. | | Deposit date: | 2005-10-26 | | Release date: | 2005-12-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis for Gas6-Axl Signalling.
Embo J., 25, 2006
|
|
1U8B
 
 | | Crystal structure of the methylated N-ADA/DNA complex | | Descriptor: | 5'-D(*AP*AP*TP*CP*TP*TP*GP*CP*GP*CP*TP*TP*T)-3', 5'-D(*TP*AP*AP*AP*TP*T)-3', 5'-D(P*AP*AP*AP*GP*CP*GP*CP*AP*AP*GP*AP*T)-3', ... | | Authors: | He, C, Hus, J.-C, Sun, L.J, Zhou, P, Norman, D.P.G, Dotsch, V, Gross, J.D, Lane, W.S, Wagner, G, Verdine, G.L. | | Deposit date: | 2004-08-05 | | Release date: | 2005-10-11 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A methylation-dependent electrostatic switch controls DNA repair and transcriptional activation by E. coli ada.
Mol.Cell, 20, 2005
|
|
1U8L
 
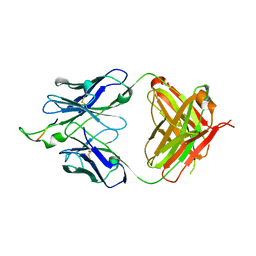 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide DLDRWAS | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-06 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
2BHM
 
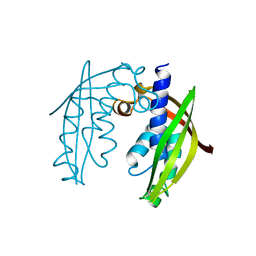 | | Crystal structure of VirB8 from Brucella suis | | Descriptor: | TYPE IV SECRETION SYSTEM PROTEIN VIRB8 | | Authors: | Bayliss, R, Baron, C, Waksman, G. | | Deposit date: | 2005-01-14 | | Release date: | 2005-03-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of Two Core Subunits of the Bacterial Type Iv Secretion System, Virb8 from Brucella Suis and Comb10 from Helicobacter Pylori
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BJ6
 
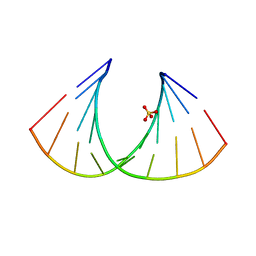 | | Crystal Structure of a decameric HNA-RNA hybrid | | Descriptor: | 5'-R(*GP*GP*CP*AP*UP*UP*AP*CP*GP*GP)-3', SULFATE ION, SYNTHETIC HNA | | Authors: | Maier, T, Przylas, I, Straeter, N, Herdewijn, P, Saenger, W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reinforced Hna Backbone Hydration in the Crystal Structure of a Decameric Hna/RNA Hybrid
J.Am.Chem.Soc., 127, 2005
|
|
2BFI
 
 | | Molecular basis for amyloid fibril formation and stability | | Descriptor: | SYNTHETIC PEPTIDE | | Authors: | Makin, O.S, Atkins, E, Sikorski, P, Johansson, J, Serpell, L.C. | | Deposit date: | 2004-12-07 | | Release date: | 2005-01-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Molecular Basis for Amyloid Fibril Formation and Stability
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2BGI
 
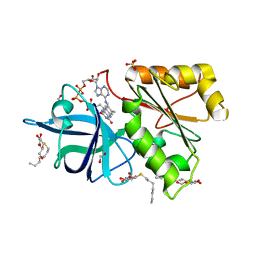 | | X-Ray Structure of the Ferredoxin-NADP(H) Reductase from Rhodobacter capsulatus complexed with three molecules of the detergent n-heptyl- beta-D-thioglucoside at 1.7 Angstroms | | Descriptor: | CARBON DIOXIDE, FERREDOXIN-NADP(H) REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Perez-Dorado, J.I, Hermoso, J.A, Nogues, I, Frago, S, Bittel, C, Mayhew, S.G, Gomez-Moreno, C, Medina, M, Cortez, N, Carrillo, N. | | Deposit date: | 2004-12-23 | | Release date: | 2005-09-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The Ferredoxin-Nadp(H) Reductase from Rhodobacter Capsulatus: Molecular Structure and Catalytic Mechanism
Biochemistry, 44, 2005
|
|
2BGU
 
 | | CRYSTAL STRUCTURE OF THE DNA MODIFYING ENZYME BETA-GLUCOSYLTRANSFERASE IN THE PRESENCE AND ABSENCE OF THE SUBSTRATE URIDINE DIPHOSPHOGLUCOSE | | Descriptor: | BETA-GLUCOSYLTRANSFERASE, URIDINE-5'-DIPHOSPHATE | | Authors: | Vrielink, A, Rueger, W, Driessen, H.P.C, Freemont, P.S. | | Deposit date: | 1994-06-09 | | Release date: | 1995-12-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the DNA modifying enzyme beta-glucosyltransferase in the presence and absence of the substrate uridine diphosphoglucose.
EMBO J., 13, 1994
|
|
1TZ2
 
 | | Crystal structure of 1-aminocyclopropane-1-carboyxlate deaminase complexed with ACC | | Descriptor: | 1-AMINOCYCLOPROPANECARBOXYLIC ACID, 1-aminocyclopropane-1-carboxylate deaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Karthikeyan, S, Zhou, Q, Zhao, Z, Kao, C.L, Tao, Z, Robinson, H, Liu, H.W, Zhang, H. | | Deposit date: | 2004-07-09 | | Release date: | 2004-11-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Analysis of Pseudomonas 1-Aminocyclopropane-1-carboxylate Deaminase Complexes: Insight into the Mechanism of a Unique Pyridoxal-5'-phosphate Dependent Cyclopropane Ring-Opening Reaction
Biochemistry, 43, 2004
|
|
