4HIF
 
 | | Ultrahigh-resolution crystal structure of Z-DNA in complex with Zn2+ ions | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), SPERMINE (FULLY PROTONATED FORM), ... | | Authors: | Drozdzal, P, Gilski, M, Kierzek, R, Lomozik, L, Jaskolski, M. | | Deposit date: | 2012-10-11 | | Release date: | 2013-06-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (0.85 Å) | | Cite: | Ultrahigh-resolution crystal structures of Z-DNA in complex with Mn(2+) and Zn(2+) ions.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
6AGF
 
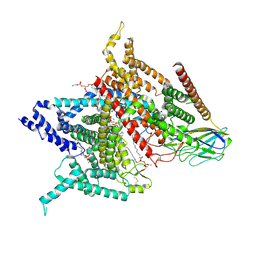 | | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta1 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, X.J, li, Z.Q, Zhou, Q, Shen, H.Z, Wu, K, Huang, X.S, Chen, J.F, Zhang, J.R, Zhu, X.C, Lei, J.L, Xiong, W, Gong, H.P, Xiao, B.L, Yan, N. | | Deposit date: | 2018-08-11 | | Release date: | 2018-10-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the human voltage-gated sodium channel Nav1.4 in complex with beta 1.
Science, 362, 2018
|
|
2FK6
 
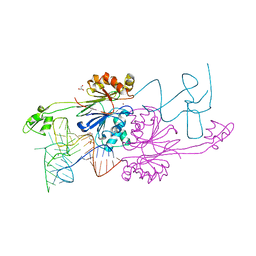 | | Crystal Structure of RNAse Z/tRNA(Thr) complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, RIBONUCLEASE Z, ... | | Authors: | Li de la Sierra-Gallay, I, Mathy, N, Pellegrini, O, Condon, C. | | Deposit date: | 2006-01-04 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the ubiquitous 3' processing enzyme RNase Z bound to transfer RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2E7Y
 
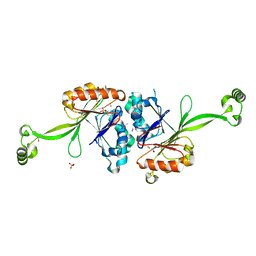 | | High resolution structure of T. maritima tRNase Z | | Descriptor: | S-1,2-PROPANEDIOL, SULFATE ION, ZINC ION, ... | | Authors: | Ishii, R, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-15 | | Release date: | 2007-09-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | The structure of the flexible arm of Thermotoga maritima tRNase Z differs from those of homologous enzymes
Acta Crystallogr.,Sect.F, 63, 2007
|
|
3CIR
 
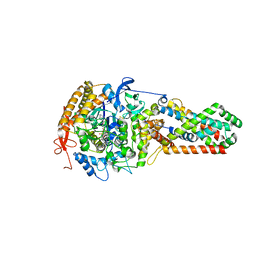 | | E. coli Quinol fumarate reductase FrdA T234A mutation | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Tomasiak, T.M, Maklashina, E, Cecchini, G, Iverson, T.M. | | Deposit date: | 2008-03-11 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | A threonine on the active site loop controls transition state formation in Escherichia coli respiratory complex II.
J.Biol.Chem., 283, 2008
|
|
1KH5
 
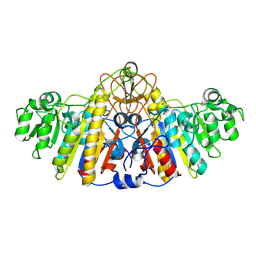 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D330N) MIMIC OF THE TRANSITION STATES WITH ALUMINIUM FLUORIDE | | Descriptor: | ALKALINE PHOSPHATASE, ALUMINUM FLUORIDE, MAGNESIUM ION, ... | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E. | | Deposit date: | 2001-11-29 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KHJ
 
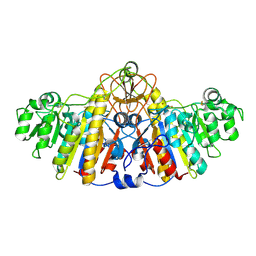 | | E. COLI ALKALINE PHOSPHATASE MUTANT (D153HD330N) MIMIC OF THE TRANSITION STATES WITH ALUMINIUM FLUORIDE | | Descriptor: | ALUMINUM FLUORIDE, Alkaline phosphatase, ZINC ION | | Authors: | Le Du, M.H, Lamoure, C, Muller, B.H, Bulgakov, O.V, Lajeunesse, E, Menez, A, Boulain, J.C. | | Deposit date: | 2001-11-30 | | Release date: | 2002-03-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Artificial evolution of an enzyme active site: structural studies of three highly active mutants of Escherichia coli alkaline phosphatase.
J.Mol.Biol., 316, 2002
|
|
1KCE
 
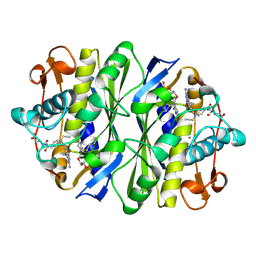 | | E. COLI THYMIDYLATE SYNTHASE MUTANT E58Q IN COMPLEX WITH CB3717 AND 2'-DEOXYURIDINE 5'-MONOPHOSPHATE (DUMP) | | Descriptor: | 10-PROPARGYL-5,8-DIDEAZAFOLIC ACID, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Sage, C.R, Rutenber, E.E, Stout, T.J, Stroud, R.M. | | Deposit date: | 1996-10-22 | | Release date: | 1997-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An essential role for water in an enzyme reaction mechanism: the crystal structure of the thymidylate synthase mutant E58Q.
Biochemistry, 35, 1996
|
|
9NLQ
 
 | |
9NL5
 
 | |
5JRD
 
 | | E. coli Hydrogenase-1 variant P508A | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Carr, S.B, Phillips, S.E.V, Armstrong, F.A, Evans, R.M, Brooke, E.J, Islam, S.T.A. | | Deposit date: | 2016-05-06 | | Release date: | 2017-03-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Importance of the Active Site "Canopy" Residues in an O2-Tolerant [NiFe]-Hydrogenase.
Biochemistry, 56, 2017
|
|
3CW5
 
 | | E. coli Initiator tRNA | | Descriptor: | Initiator tRNA | | Authors: | Barraud, P, Schmitt, E, Mechulam, Y, Dardel, F, Tisne, C. | | Deposit date: | 2008-04-21 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis.
Nucleic Acids Res., 36, 2008
|
|
8RND
 
 | | Cathepsin S in complex with NNPI-C10 inhibitor | | Descriptor: | 1,2-ETHANEDIOL, Cathepsin S, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Petruzzella, A, Lau, K, Pojer, F, Oricchio, E. | | Deposit date: | 2024-01-09 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Antibody-peptide conjugates deliver covalent inhibitors blocking oncogenic cathepsins.
Nat.Chem.Biol., 20, 2024
|
|
9NLB
 
 | |
9NLJ
 
 | | E.coli Initiation complex with Uup-EQ2 | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Singh, S, Hunt, J.F. | | Deposit date: | 2025-03-03 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | E.coli Initiation complex with Uup-EQ2
To Be Published
|
|
9NLS
 
 | |
9NKL
 
 | | E. coli 70S initiation complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Singh, S, Hunt, J.F. | | Deposit date: | 2025-03-01 | | Release date: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | E. coli 70S initiation complex
To Be Published
|
|
9NJF
 
 | |
9NJV
 
 | |
1KMK
 
 | |
1KMJ
 
 | |
3D27
 
 | | E. coli methionine aminopeptidase with Fe inhibitor W29 | | Descriptor: | 4-(3-ethylthiophen-2-yl)benzene-1,2-diol, MANGANESE (II) ION, Methionine aminopeptidase | | Authors: | Ye, Q.Z, Chai, S, He, H.Z. | | Deposit date: | 2008-05-07 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of inhibitors of Escherichia coli methionine aminopeptidase with the Fe(II)-form selectivity and antibacterial activity.
J.Med.Chem., 51, 2008
|
|
8RJ6
 
 | | E. coli adenylate kinase in complex with ATP and AMP and Mg2+ as a result of enzymatic AP4A hydrolysis. | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Tischlik, S, Ronge, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2023-12-20 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Magnesium induced structural reorganization in the active site of adenylate kinase.
Sci Adv, 10, 2024
|
|
8RJ4
 
 | | E. coli adenylate kinase in complex with two ADP molecules and Mg2+ as a result of enzymatic AP4A hydrolysis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase, CHLORIDE ION, ... | | Authors: | Tischlik, S, Ronge, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2023-12-20 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Magnesium induced structural reorganization in the active site of adenylate kinase.
Sci Adv, 10, 2024
|
|
8RJ9
 
 | | E. coli adenylate kinase Asp84Ala variant in complex with two ADP molecules as a result of enzymatic AP4A hydrolysis. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Adenylate kinase | | Authors: | Tischlik, S, Ronge, P, Wolf-Watz, M, Sauer-Eriksson, A.E. | | Deposit date: | 2023-12-20 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Magnesium induced structural reorganization in the active site of adenylate kinase.
Sci Adv, 10, 2024
|
|
