2DU6
 
 | |
2DTJ
 
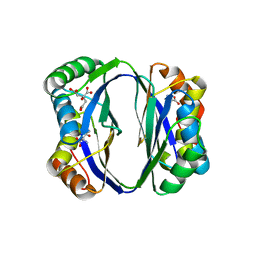 | | Crystal structure of regulatory subunit of aspartate kinase from Corynebacterium glutamicum | | Descriptor: | Aspartokinase, CITRIC ACID, THREONINE | | Authors: | Yoshida, A, Tomita, T, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2006-07-12 | | Release date: | 2007-05-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structural Insight into Concerted Inhibition of alpha(2)beta(2)-Type Aspartate Kinase from Corynebacterium glutamicum
J.Mol.Biol., 368, 2007
|
|
1U05
 
 | |
1U0Z
 
 | |
1BY8
 
 | | THE CRYSTAL STRUCTURE OF HUMAN PROCATHEPSIN K | | Descriptor: | PROTEIN (PROCATHEPSIN K) | | Authors: | Lalonde, J.M, Zhao, B, Smith, W.W, Janson, C.A, Desjarlais, R.L, Tomaszek, T.A, Carr, T.J, Thompson, S.K, Yamashita, D.S, Veber, D.F, Abdel-Mequid, S.S. | | Deposit date: | 1998-10-27 | | Release date: | 1999-10-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of human procathepsin K.
Biochemistry, 38, 1999
|
|
1W59
 
 | | FtsZ dimer, empty (M. jannaschii) | | Descriptor: | CELL DIVISION PROTEIN FTSZ HOMOLOG 1, SULFATE ION | | Authors: | Oliva, M.A, Cordell, S.C, Lowe, J. | | Deposit date: | 2004-08-06 | | Release date: | 2004-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural Insights Into Ftsz Protofilament Formation
Nat.Struct.Mol.Biol., 11, 2004
|
|
1W35
 
 | | FERREDOXIN-NADP+ REDUCTASE (MUTATION: Y 303 W) | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Perez-Dorado, I, Medina, M, Julvez, M.M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2004-07-13 | | Release date: | 2005-07-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | C-Terminal Tyrosine of Ferredoxin-Nadp(+) Reductase in Hydride Transfer Processes with Nad(P)(+)/H.
Biochemistry, 44, 2005
|
|
1W34
 
 | | FERREDOXIN-NADP REDUCTASE (MUTATION: Y 303 S) | | Descriptor: | FERREDOXIN-NADP REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Hermoso, J.A, Perez-Dorado, I, Medina, M, Julvez, M.M, Sanz-Aparicio, J, Gomez-Moreno, C. | | Deposit date: | 2004-07-13 | | Release date: | 2005-10-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | C-Terminal Tyrosine of Ferredoxin-Nadp(+) Reductase in Hydride Transfer Processes with Nad(P)(+)/H.
Biochemistry, 44, 2005
|
|
1W6Y
 
 | | crystal structure of a mutant W92A in ketosteroid isomerase (KSI) from Pseudomonas putida biotype B | | Descriptor: | BETA-MERCAPTOETHANOL, EQUILENIN, STEROID DELTA-ISOMERASE | | Authors: | Yun, Y.S, Nam, G.H, Kim, Y.-G, Oh, B.-H, Choi, K.Y. | | Deposit date: | 2004-08-25 | | Release date: | 2005-04-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Small Exterior Hydrophobic Cluster Contributes to Conformational Stability and Steroid Binding in Ketosteroid Isomerase from Pseudomonas Putida Biotype B
FEBS J., 272, 2005
|
|
1BOH
 
 | | SULFUR-SUBSTITUTED RHODANESE (ORTHORHOMBIC FORM) | | Descriptor: | RHODANESE | | Authors: | Gliubich, F, Berni, R, Cianci, M, Trevino, R.J, Horowitz, P.M, Zanotti, G. | | Deposit date: | 1998-08-04 | | Release date: | 1999-04-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | NH2-terminal sequence truncation decreases the stability of bovine rhodanese, minimally perturbs its crystal structure, and enhances interaction with GroEL under native conditions.
J.Biol.Chem., 274, 1999
|
|
2DFE
 
 | | Crystal structure of Tk-RNase HII(1-200)-C | | Descriptor: | Ribonuclease HII | | Authors: | Katagiri, Y, Takano, K, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2006-03-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational contagion in a protein: Structural properties of a chameleon sequence
Proteins, 68, 2007
|
|
1W99
 
 | |
2DFF
 
 | | Crystal structure of Tk-RNase HII(1-204)-C | | Descriptor: | Ribonuclease HII | | Authors: | Katagiri, Y, Takano, K, Chon, H, Matsumura, H, Koga, Y, Kanaya, S. | | Deposit date: | 2006-03-01 | | Release date: | 2007-03-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational contagion in a protein: Structural properties of a chameleon sequence
Proteins, 68, 2007
|
|
1WB7
 
 | | Iron Superoxide Dismutase (Fe-SOD) From The Hyperthermophile Sulfolobus Solfataricus. Crystal Structure of the Y41F mutant. | | Descriptor: | FE (III) ION, SUPEROXIDE DISMUTASE [FE] | | Authors: | Gogliettino, M.A, Tanfani, F, Scire, A, Ursby, T, Adinolfi, B.S, Cacciamani, T, De Vendittis, E. | | Deposit date: | 2004-10-31 | | Release date: | 2004-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | The Role of Tyr41 and His155 in the Functional Properties of Superoxide Dismutase from the Archaeon Sulfolobus Solfataricus
Biochemistry, 43, 2004
|
|
2DKA
 
 | | Crystal structure of N-acetylglucosamine-phosphate mutase, a member of the alpha-D-phosphohexomutase superfamily, in the apo-form | | Descriptor: | Phosphoacetylglucosamine mutase | | Authors: | Nishitani, Y, Maruyama, D, Nonaka, T, Kita, A, Fukami, T.A, Mio, T, Yamada-Okabe, H, Yamada-Okabe, T, Miki, K. | | Deposit date: | 2006-04-07 | | Release date: | 2006-05-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structures of N-Acetylglucosamine-phosphate Mutase, a Member of the {alpha}-D-Phosphohexomutase Superfamily, and Its Substrate and Product Complexes.
J.Biol.Chem., 281, 2006
|
|
1W5F
 
 | | FtsZ, T7 mutated, domain swapped (T. maritima) | | Descriptor: | CELL DIVISION PROTEIN FTSZ, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER | | Authors: | Oliva, M.A, Cordell, S.C, Lowe, J. | | Deposit date: | 2004-08-06 | | Release date: | 2004-12-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights Into Ftsz Protofilament Formation
Nat.Struct.Mol.Biol., 11, 2004
|
|
1U1S
 
 | | Hfq protein from Pseudomonas aeruginosa. Low-salt crystals | | Descriptor: | Hfq protein | | Authors: | Nikulin, A.D, Stolboushkina, E.A, Perederina, I, Vassilieva, I.M, Blaesi, U, Moll, I, Kachalova, G, Vassylyev, D, Yokoyama, S, Garber, M, Nikonov, S.V, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-16 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Pseudomonas aeruginosa Hfq protein.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1U91
 
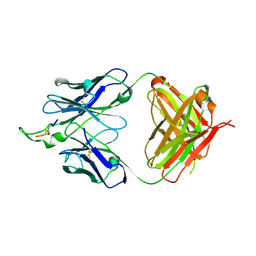 | | Crystal structure of the HIV-1 Cross Neutralizing Monoclonal Antibody 2F5 in complex with gp41 Peptide Analog ENDKW-[Dap]-S (cyclic) | | Descriptor: | ANTIBODY 2F5 (HEAVY CHAIN), ANTIBODY 2F5 (LIGHT CHAIN), GP41 PEPTIDE ANALOG | | Authors: | Bryson, S, Julien, J.-P, Hynes, R.C, Pai, E.F. | | Deposit date: | 2004-08-09 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystallographic definition of the epitope promiscuity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5: vaccine design implications
J.Virol., 83, 2009
|
|
1W4P
 
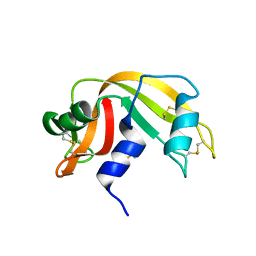 | | Binding of Nonnatural 3'-Nucleotides to Ribonuclease A | | Descriptor: | 2'-DEOXYURIDINE 3'-MONOPHOSPHATE, PANCREATIC RIBONUCLEASE A | | Authors: | Jenkins, C.L, Thiyagarajan, N, Sweeney, R.Y, Guy, M.P, Kelemen, B.R, Acharya, K.R, Raines, R.T. | | Deposit date: | 2004-07-27 | | Release date: | 2005-02-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Binding of Non-Natural 3'-Nucleotides to Ribonuclease A
FEBS J., 272, 2005
|
|
1FNT
 
 | | CRYSTAL STRUCTURE OF THE 20S PROTEASOME FROM YEAST IN COMPLEX WITH THE PROTEASOME ACTIVATOR PA26 FROM TRYPANOSOME BRUCEI AT 3.2 ANGSTROMS RESOLUTION | | Descriptor: | MAGNESIUM ION, PROTEASOME ACTIVATOR PROTEIN PA26, PROTEASOME COMPONENT C1, ... | | Authors: | Whitby, F.G, Masters, E, Kramer, L, Knowlton, J.R, Yao, Y, Wang, C.C, Hill, C.P. | | Deposit date: | 2000-08-23 | | Release date: | 2001-04-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for the activation of 20S proteasomes by 11S regulators.
Nature, 408, 2000
|
|
1UAM
 
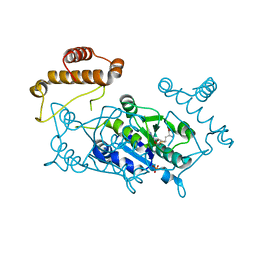 | | Crystal structure of tRNA(m1G37)methyltransferase: Insight into tRNA recognition | | Descriptor: | PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Guanine-N(1)-)-methyltransferase | | Authors: | Ahn, H.J, Kim, H.-W, Yoon, H.-J, Lee, B.I, Suh, S.W, Yang, J.K. | | Deposit date: | 2003-03-11 | | Release date: | 2003-06-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of tRNA(m(1)G37)methyltransferase: insights into tRNA recognition
EMBO J., 22, 2003
|
|
1W7W
 
 | |
1U2Q
 
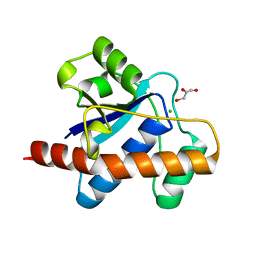 | | Crystal structure of Mycobacterium tuberculosis Low Molecular Weight Protein Tyrosine Phosphatase (MPtpA) at 2.5A resolution with glycerol in the active site | | Descriptor: | CHLORIDE ION, GLYCEROL, low molecular weight protein-tyrosine-phosphatase | | Authors: | Madhurantakam, C, Rajakumara, E, Mazumdar, P.A, Saha, B, Mitra, D, Wiker, H.G, Sankaranarayanan, R, Das, A.K. | | Deposit date: | 2004-07-20 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Low-Molecular-Weight Protein Tyrosine Phosphatase from Mycobacterium tuberculosis at 1.9-A Resolution
J.Bacteriol., 187, 2005
|
|
1W8A
 
 | |
1F8S
 
 | | CRYSTAL STRUCTURE OF L-AMINO ACID OXIDASE FROM CALLOSELASMA RHODOSTOMA, COMPLEXED WITH THREE MOLECULES OF O-AMINOBENZOATE. | | Descriptor: | 2-AMINOBENZOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Pawelek, P.D, Cheah, J, Coulombe, R, Macheroux, P, Ghisla, S, Vrielink, A. | | Deposit date: | 2000-07-04 | | Release date: | 2000-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of L-amino acid oxidase reveals the substrate trajectory into an enantiomerically conserved active site.
EMBO J., 19, 2000
|
|
