8JZJ
 
 | | E.coli Glyceraldehyde-3-phosphate dehydrogenase structure under cryoprotect condition of ammonium sulfate | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Jang, K, Hlaing, S.H.S, Kim, H.G, Kim, N, Choe, H.W, Kim, Y.J. | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | E.coli Glyceraldehyde-3-phosphate dehydrogenase structure under cryoprotect condition of ammonium sulfate
To Be Published
|
|
7K4W
 
 | | Crystal structure of Kemp Eliminase HG3.17 in the inactive state | | Descriptor: | CALCIUM ION, Endo-1,4-beta-xylanase | | Authors: | Padua, R.A.P, Otten, R, Bunzel, A, Nguyen, V, Pitsawong, W, Patterson, M, Sui, S, Perry, S.L, Cohen, A.E, Hilvert, D, Kern, D. | | Deposit date: | 2020-09-16 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | How directed evolution reshapes the energy landscape in an enzyme to boost catalysis.
Science, 370, 2020
|
|
6W8E
 
 | |
1RF8
 
 | | Solution structure of the yeast translation initiation factor eIF4E in complex with m7GDP and eIF4GI residues 393 to 490 | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, Eukaryotic initiation factor 4F subunit p150, Eukaryotic translation initiation factor 4E, ... | | Authors: | Gross, J.D, Moerke, N.J, von der Haar, T, Lugovskoy, A.A, Sachs, A.B, McCarthy, J.E.G, Wagner, G. | | Deposit date: | 2003-11-07 | | Release date: | 2003-12-23 | | Last modified: | 2024-03-06 | | Method: | SOLUTION NMR | | Cite: | Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E.
Cell(Cambridge,Mass.), 115, 2003
|
|
5UJK
 
 | |
5UJU
 
 | |
5YNH
 
 | | Crystal structure of Pullulanase from Klebsiella pneumoniae complex at 10 mM gamma-cyclodextrin | | Descriptor: | CALCIUM ION, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), GLYCEROL, ... | | Authors: | Saka, N, Iwamoto, H, Takahashi, N, Mizutani, K, Mikami, B. | | Deposit date: | 2017-10-24 | | Release date: | 2018-10-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Elucidation of the mechanism of interaction between Klebsiella pneumoniae pullulanase and cyclodextrin
Acta Crystallogr D Struct Biol, 74, 2018
|
|
8RT2
 
 | | Hen Egg White Lysozyme Crystallized with Bioassembler on Earth | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, CHLORIDE ION, GADOLINIUM ATOM, ... | | Authors: | MacCarthy, C, Koudan, E.V, Petrov, S.V, Levin, A.A, Khesuani, Y.D, Borshchevskiy, V. | | Deposit date: | 2024-01-25 | | Release date: | 2024-03-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | Hen Egg White Lysozyme Crystallized with Bioassembler on Earth
To Be Published
|
|
3BS1
 
 | | Structure of the Staphylococcus aureus AgrA LytTR Domain Bound to DNA Reveals a Beta Fold with a Novel Mode of Binding | | Descriptor: | Accessory gene regulator protein A, DNA (5'-D(*DAP*DAP*(BRU)P*DAP*DCP*DTP*DTP*DAP*DAP*DCP*DTP*DGP*DTP*DTP*DAP*DA)-3'), DNA (5'-D(*DTP*DTP*DTP*DAP*DAP*DCP*DAP*DGP*DTP*DTP*DAP*DAP*DGP*(BRU)P*DAP*DT)-3'), ... | | Authors: | Sidote, D.J, Barbieri, C, Wu, T, Stock, A.M. | | Deposit date: | 2007-12-21 | | Release date: | 2008-04-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the Staphylococcus aureus AgrA LytTR Domain Bound to DNA Reveals a Beta Fold with an Unusual Mode of Binding.
Structure, 16, 2008
|
|
5I04
 
 | | Crystal structure of the orphan region of human endoglin/CD105 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Maltose-binding periplasmic protein,Endoglin, TRIETHYLENE GLYCOL, ... | | Authors: | Saito, T, Bokhove, M, de Sanctis, D, Jovine, L. | | Deposit date: | 2016-02-03 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1.
Cell Rep, 19, 2017
|
|
1RIO
 
 | | Structure of bacteriophage lambda cI-NTD in complex with sigma-region4 of Thermus aquaticus bound to DNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 27-MER, CALCIUM ION, ... | | Authors: | Jain, D, Nickels, B.E, Sun, L, Hochschild, A, Darst, S.A. | | Deposit date: | 2003-11-17 | | Release date: | 2004-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a ternary transcription activation complex.
Mol.Cell, 13, 2004
|
|
7JWK
 
 | |
8RGP
 
 | | Closed Complex I from murine brain | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Vercellino, I, Sazanov, L.A. | | Deposit date: | 2023-12-14 | | Release date: | 2024-04-24 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | SCAF1 drives the compositional diversity of mammalian respirasomes.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8QZF
 
 | | Heme-domain BM3 mutant T268E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Opperman, D.J, Ebrecht, A.C, Aschenbrenner, J.C. | | Deposit date: | 2023-10-27 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Revisiting strategies and their combinatorial effect for introducing peroxygenase activity in CYP102A1 (P450BM3)
Mol Catal, 557, 2024
|
|
5V36
 
 | | 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-03-06 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure of Glutathione Reductase from Streptococcus mutans UA159 in Complex with FAD.
To Be Published
|
|
7XYG
 
 | |
5YQR
 
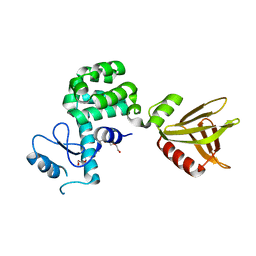 | | Crystal structure of the PH-like domain of Lam6 | | Descriptor: | Endolysin/Membrane-anchored lipid-binding protein LAM6 fusion protein, NONAETHYLENE GLYCOL | | Authors: | Tong, J, Im, Y.J. | | Deposit date: | 2017-11-07 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.402 Å) | | Cite: | Structural basis of sterol recognition and nonvesicular transport by lipid transfer proteins anchored at membrane contact sites
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6W1V
 
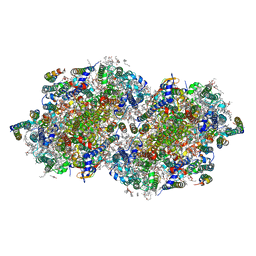 | | RT XFEL structure of the two-flash state of Photosystem II (2F, S3-rich) at 2.09 Angstrom resolution | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Ibrahim, M, Fransson, T, Chatterjee, R, Cheah, M.H, Hussein, R, Lassalle, L, Sutherlin, K.D, Young, I.D, Fuller, F.D, Gul, S, Kim, I.-S, Simon, P.S, de Lichtenberg, C, Chernev, P, Bogacz, I, Pham, C, Orville, A.M, Saichek, N, Northen, T.R, Batyuk, A, Carbajo, S, Alonso-Mori, R, Tono, K, Owada, S, Bhowmick, A, Bolotovski, R, Mendez, D, Moriarty, N.W, Holton, J.M, Dobbek, H, Brewster, A.S, Adams, P.D, Sauter, N.K, Bergmann, U, Zouni, A, Messinger, J, Kern, J, Yachandra, V.K, Yano, J. | | Deposit date: | 2020-03-04 | | Release date: | 2020-06-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Untangling the sequence of events during the S2→ S3transition in photosystem II and implications for the water oxidation mechanism.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5B12
 
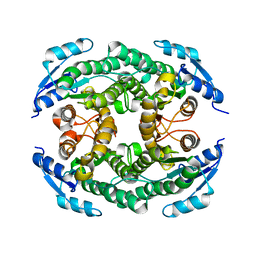 | | Crystal structure of the B-type halohydrin hydrogen-halide-lyase mutant F71W/Q125T/D199H from Corynebacterium sp. N-1074 | | Descriptor: | CHLORIDE ION, Halohydrin epoxidase B | | Authors: | Watanabe, F, Yu, F, Ohtaki, A, Yamanaka, Y, Noguchi, K, Odaka, M, Yohda, M. | | Deposit date: | 2015-11-17 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.721 Å) | | Cite: | Improvement of enantioselectivity of the B-type halohydrin hydrogen-halide-lyase from Corynebacterium sp. N-1074
J.Biosci.Bioeng., 122, 2016
|
|
8QK2
 
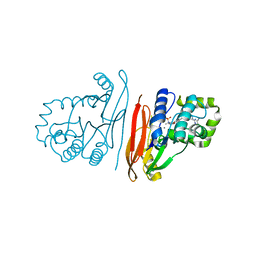 | | Structure of K.pneumoniae LpxH in complex with EBL-3339 | | Descriptor: | MANGANESE (II) ION, UDP-2,3-diacylglucosamine hydrolase, ~{N}-[4-[4-(4-cyano-6-methyl-pyrimidin-2-yl)piperazin-1-yl]sulfonylphenyl]-2-[methyl(methylsulfonyl)amino]benzamide | | Authors: | Sooriyaarachchi, S, Bergfors, T, Jones, T.A, Mowbray, S.L. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibiotic class with potent in vivo activity targeting lipopolysaccharide synthesis in Gram-negative bacteria.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
1KDT
 
 | | CYTIDINE MONOPHOSPHATE KINASE FROM E.COLI IN COMPLEX WITH 2',3'-DIDEOXY-CYTIDINE MONOPHOSPHATE | | Descriptor: | 2',3'-DIDEOXYCYTIDINE-5'-MONOPHOSPHATE, CYTIDYLATE KINASE, SULFATE ION | | Authors: | Bertrand, T, Briozzo, P, Assairi, L, Ofiteru, A, Bucurenci, N, Munier-Lehmann, H, Golinelli-Pimpaneau, B, Barzu, O, Gilles, A.M. | | Deposit date: | 2001-11-13 | | Release date: | 2002-01-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Sugar specificity of bacterial CMP kinases as revealed by crystal structures and mutagenesis of Escherichia coli enzyme.
J.Mol.Biol., 315, 2002
|
|
5N48
 
 | |
5B2F
 
 | | N,N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii complexed with its inhibitor MPG (phosphate-containing condition) | | Descriptor: | 2-deoxy-2-{[(S)-hydroxy(methyl)phosphoryl]amino}-beta-D-glucopyranose, Putative uncharacterized protein PH0499, ZINC ION | | Authors: | Nakamura, T, Niiyama, M, Ida, K, Uegaki, K. | | Deposit date: | 2016-01-15 | | Release date: | 2016-08-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Substrate recognition of N,N'-diacetylchitobiose deacetylase from Pyrococcus horikoshii
J.Struct.Biol., 195, 2016
|
|
5IF6
 
 | |
5YQS
 
 | | Isoprimeverose-producing enzyme from Aspergillus oryzae in complex with isoprimeverose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yaoi, K, Matsuzawa, T, Watanabe, M, Nakamichi, Y. | | Deposit date: | 2017-11-07 | | Release date: | 2018-11-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and substrate recognition mechanism of Aspergillus oryzae isoprimeverose-producing enzyme.
J.Struct.Biol., 205, 2019
|
|
