7VR6
 
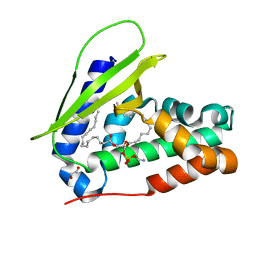 | | Crystal structure of MlaC from Escherichia coli in quasi-open state | | Descriptor: | 1,2-ETHANEDIOL, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, Intermembrane phospholipid transport system binding protein MlaC | | Authors: | Dutta, A, Kanaujia, S.P. | | Deposit date: | 2021-10-21 | | Release date: | 2022-09-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MlaC belongs to a unique class of non-canonical substrate-binding proteins and follows a novel phospholipid-binding mechanism.
J.Struct.Biol., 214, 2022
|
|
4ACF
 
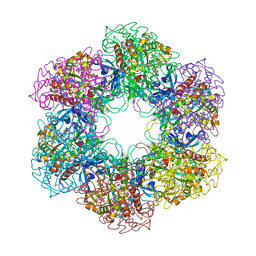 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE IN COMPLEX WITH IMIDAZOPYRIDINE INHIBITOR ((4-(6-BROMO-3-(BUTYLAMINO)IMIDAZO(1,2-A)PYRIDIN-2-YL)PHENOXY) ACETIC ACID) AND L-METHIONINE-S-SULFOXIMINE PHOSPHATE. | | Descriptor: | CHLORIDE ION, GLUTAMINE SYNTHETASE 1, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | Authors: | Nilsson, M.T, Mowbray, S.L. | | Deposit date: | 2011-12-15 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, Biological Evaluation and X-Ray Crystallographic Studies of Imidazo(1,2-A)Pyridine-Based Mycobacterium Tuberculosis Glutamine Synthetase Inhibitors
Medchemcomm, 3, 2012
|
|
8BYI
 
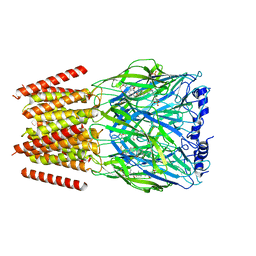 | | Alvinella pompejana nicotinic acetylcholine receptor Alpo4 in complex with CHAPS(Alpo4_CHAPS) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Acetylcholine receptor, ... | | Authors: | De Gieter, S, Efremov, R.G, Ulens, C. | | Deposit date: | 2022-12-13 | | Release date: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Sterol derivative binding to the orthosteric site causes conformational changes in an invertebrate Cys-loop receptor.
Elife, 12, 2023
|
|
7SV3
 
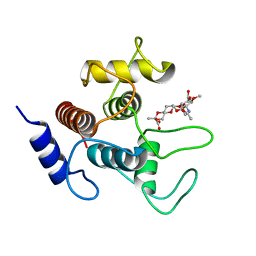 | | Crystal structure of SpaA-SLH in complex with 4,6-Pyr-beta-D-ManNAc-(1->4)-beta-D-GlcNAcOMe | | Descriptor: | Surface (S-) layer glycoprotein, methyl 2-acetamido-4-O-{2-acetamido-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl}-2-deoxy-beta-D-glucopyranoside | | Authors: | Legg, M.S.G, Evans, S.V. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The S-layer homology domains of Paenibacillus alvei surface protein SpaA bind to cell wall polysaccharide through the terminal monosaccharide residue.
J.Biol.Chem., 298, 2022
|
|
7SV4
 
 | | Crystal structure of SpaA-SLH in complex with 4,6-Pyr-beta-D-ManNAc-(1->4)-beta-D-GlcNAc-(1->3)-4,6-Pyr-beta-D-ManNAcOMe | | Descriptor: | GLYCEROL, Surface (S-) layer glycoprotein, methyl 2-acetamido-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl-(1->4)-2-acetamido-2-deoxy-beta-D-glucopyranosyl-(1->3)-2-acetamido-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranoside | | Authors: | Legg, M.S.G, Evans, S.V. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The S-layer homology domains of Paenibacillus alvei surface protein SpaA bind to cell wall polysaccharide through the terminal monosaccharide residue.
J.Biol.Chem., 298, 2022
|
|
3QP6
 
 | | Crystal structure of CviR (Chromobacterium violaceum 12472) bound to C6-HSL | | Descriptor: | CviR transcriptional regulator, N-[(3S)-2-oxotetrahydrofuran-3-yl]hexanamide | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
2HGS
 
 | | HUMAN GLUTATHIONE SYNTHETASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLUTATHIONE, MAGNESIUM ION, ... | | Authors: | Polekhina, G, Board, P, Rossjohn, J, Parker, M.W. | | Deposit date: | 1999-01-04 | | Release date: | 1999-06-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis of glutathione synthetase deficiency and a rare gene permutation event.
EMBO J., 18, 1999
|
|
3Q47
 
 | |
3Q49
 
 | |
3ULU
 
 | | Structure of quaternary complex of human TLR3ecd with three Fabs (Form1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1068 heavy chain, ... | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
7BY6
 
 | | Plasmodium vivax cytoplasmic Phenylalanyl-tRNA synthetase in complex with BRD1389 | | Descriptor: | (3S,4R,8R,9R,10S)-N-(4-cyclopropyloxyphenyl)-10-(methoxymethyl)-3,4-bis(oxidanyl)-9-[4-(2-phenylethynyl)phenyl]-1,6-diazabicyclo[6.2.0]decane-6-carboxamide, MAGNESIUM ION, Phenylalanyl-tRNA synthetase beta chain, ... | | Authors: | Malhotra, N, Manmohan, S, Harlos, K, Melillo, B, Schreiber, S.L, Manickam, Y, Sharma, S. | | Deposit date: | 2020-04-21 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.997 Å) | | Cite: | Structural basis of malaria parasite phenylalanine tRNA-synthetase inhibition by bicyclic azetidines.
Nat Commun, 12, 2021
|
|
3ULV
 
 | | Structure of quaternary complex of human TLR3ecd with three Fabs (Form2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab1068 heavy chain, ... | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.522 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
3MHD
 
 | | Crystal structure of DCR3 | | Descriptor: | Tumor necrosis factor receptor superfamily member 6B | | Authors: | Zhan, C, Patskovsky, Y, Yan, Q, Li, Z, Ramagopal, U.A, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2010-04-07 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Structure, 19, 2011
|
|
2AQD
 
 | | cytochrome c peroxidase (CCP) in complex with 2,5-diaminopyridine | | Descriptor: | Cytochrome c peroxidase, mitochondrial, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Brenk, R, Vetter, S.W, Boyce, S.E, Goodin, D.B, Shoichet, B.K. | | Deposit date: | 2005-08-17 | | Release date: | 2006-04-11 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Probing molecular docking in a charged model binding site.
J.Mol.Biol., 357, 2006
|
|
7SV6
 
 | |
7SV5
 
 | | Crystal structure of SpaA-SLH/G109A in complex with 4,6-Pyr-beta-D-ManNAc-(1->4)-beta-D-GlcNAcOMe | | Descriptor: | GLYCEROL, Surface (S-) layer glycoprotein, methyl 2-acetamido-4-O-{2-acetamido-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranosyl}-2-deoxy-beta-D-glucopyranoside | | Authors: | Legg, M.S.G, Evans, S.V. | | Deposit date: | 2021-11-18 | | Release date: | 2022-03-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | The S-layer homology domains of Paenibacillus alvei surface protein SpaA bind to cell wall polysaccharide through the terminal monosaccharide residue.
J.Biol.Chem., 298, 2022
|
|
8QEZ
 
 | | Crystal structure of the AMPA receptor GluA2-L504Y-N775S ligand binding domain in complex with L-glutamate and positive allosteric modulator BPAM395 at 1.55A resolution | | Descriptor: | 6-chloranyl-4-cyclopropyl-2,3-dihydrothieno[3,2-e][1,2,4]thiadiazine 1,1-dioxide, ACETATE ION, CACODYLATE ION, ... | | Authors: | Dorosz, J, Laulumaa, S, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Exploring thienothiadiazine dioxides as isosteric analogues of benzo- and pyridothiadiazine dioxides in the search of new AMPA and kainate receptor positive allosteric modulators.
Eur.J.Med.Chem., 264, 2023
|
|
3ULS
 
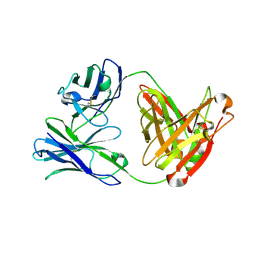 | | Crystal structure of Fab12 | | Descriptor: | Fab12 heavy chain, Fab12 light chain | | Authors: | Luo, J, Gilliland, G.L, Obmolova, O, Malia, T, Teplyakov, A. | | Deposit date: | 2011-11-11 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.495 Å) | | Cite: | Lateral Clustering of TLR3:dsRNA Signaling Units Revealed by TLR3ecd:3Fabs Quaternary Structure.
J.Mol.Biol., 421, 2012
|
|
3M0K
 
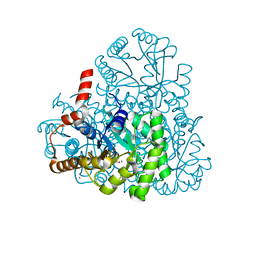 | |
8SPP
 
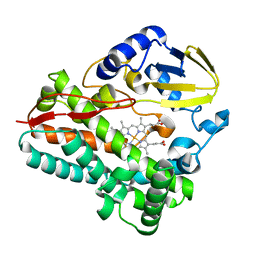 | |
3MXE
 
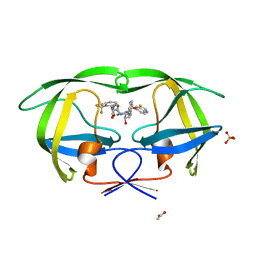 | | Crystal structure of HIV-1 protease inhibitor, KC32 complexed with wild-type protease | | Descriptor: | (5S)-N-{(1S,2R)-3-[(1,3-benzothiazol-6-ylsulfonyl)(2-methylpropyl)amino]-1-benzyl-2-hydroxypropyl}-2-oxo-3-[2-(trifluoromethyl)phenyl]-1,3-oxazolidine-5-carboxamide, ACETATE ION, HIV-1 protease, ... | | Authors: | Nalam, M.N.L, Schiffer, C.A. | | Deposit date: | 2010-05-07 | | Release date: | 2010-11-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Based Design, Synthesis, and Structure-Activity Relationship Studies of HIV-1 Protease Inhibitors Incorporating Phenyloxazolidinones.
J.Med.Chem., 53, 2010
|
|
3QP5
 
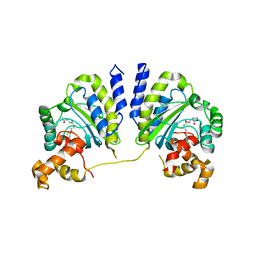 | | Crystal structure of CviR bound to antagonist chlorolactone (CL) | | Descriptor: | 4-(4-chlorophenoxy)-N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, CviR transcriptional regulator | | Authors: | Chen, G, Swem, L, Swem, D, Stauff, D, O'Loughlin, C, Jeffrey, P, Bassler, B, Hughson, F. | | Deposit date: | 2011-02-11 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.249 Å) | | Cite: | A strategy for antagonizing quorum sensing.
Mol.Cell, 42, 2011
|
|
7RYZ
 
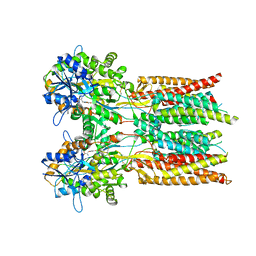 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit GSG1L bound to agonist quisqualate | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-26 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (4.15 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
8AJO
 
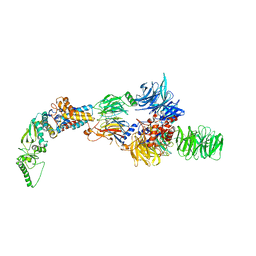 | | Negative-stain electron microscopy structure of DDB1-DCAF12-CCT5 | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | Authors: | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-09 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (30.6 Å) | | Cite: | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
7RZ5
 
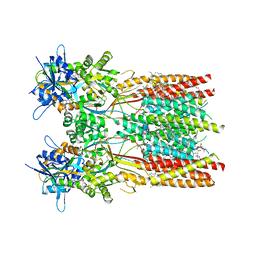 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit TARP gamma-5 bound to competitive antagonist ZK 200775 | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Glutamate receptor 2, {[7-morpholin-4-yl-2,3-dioxo-6-(trifluoromethyl)-3,4-dihydroquinoxalin-1(2H)-yl]methyl}phosphonic acid | | Authors: | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | Deposit date: | 2021-08-27 | | Release date: | 2021-10-27 | | Last modified: | 2022-02-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
