5AYK
 
 | | Crystal structure of ERdj5 form I | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CHLORIDE ION, DnaJ homolog subfamily C member 10 | | Authors: | Watanabe, S, Maegawa, K, Inaba, K. | | Deposit date: | 2015-08-22 | | Release date: | 2017-02-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Highly dynamic nature of ERdj5 is essential for enhancement of the ER associated degradation
To Be Published
|
|
5CW4
 
 | | Structure of CfBRCC36-CfKIAA0157 complex (Selenium Edge) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, GLYCEROL, Protein FAM175B, ... | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.543 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
1TFI
 
 | | A NOVEL ZN FINGER MOTIF IN THE BASAL TRANSCRIPTIONAL MACHINERY: THREE-DIMENSIONAL NMR STUDIES OF THE NUCLEIC-ACID BINDING DOMAIN OF TRANSCRIPTIONAL ELONGATION FACTOR TFIIS | | Descriptor: | TRANSCRIPTIONAL ELONGATION FACTOR SII, ZINC ION | | Authors: | Qian, X, Gozani, S, Yoon, H.S, Jeon, C.J, Agarwal, K, Weiss, M.A. | | Deposit date: | 1993-04-27 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Novel zinc finger motif in the basal transcriptional machinery: three-dimensional NMR studies of the nucleic acid binding domain of transcriptional elongation factor TFIIS.
Biochemistry, 32, 1993
|
|
1MR1
 
 | | Crystal Structure of a Smad4-Ski Complex | | Descriptor: | Mothers against decapentaplegic homolog 4, Ski oncogene, ZINC ION | | Authors: | Wu, J.-W, Krawitz, A.R, Chai, J, Li, W, Zhang, F, Luo, K, Shi, Y. | | Deposit date: | 2002-09-17 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Mechanism of Smad4 Recognition by the Nuclear Oncoprotein Ski: Insights on Ski-mediated Repression of TGF-beta Signaling
Cell(Cambridge,Mass.), 111, 2002
|
|
5CW3
 
 | | Structure of CfBRCC36-CfKIAA0157 complex (Zn Edge) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, Protein FAM175B, ZINC ION | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
5CW5
 
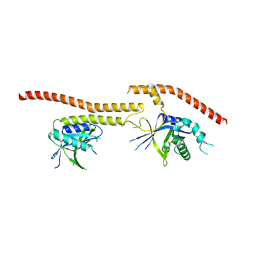 | | Structure of CfBRCC36-CfKIAA0157 complex (QSQ mutant) | | Descriptor: | BRCA1/BRCA2-containing complex subunit 3, Protein FAM175B | | Authors: | Zeqiraj, E. | | Deposit date: | 2015-07-27 | | Release date: | 2015-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.736 Å) | | Cite: | Higher-Order Assembly of BRCC36-KIAA0157 Is Required for DUB Activity and Biological Function.
Mol.Cell, 59, 2015
|
|
1A8V
 
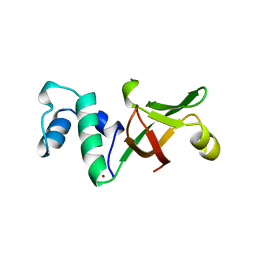 | |
4B33
 
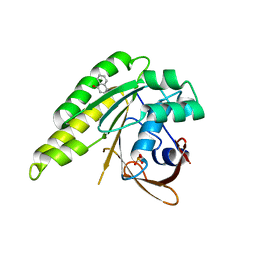 | | Humanised monomeric RadA in complex with napht-2-ol | | Descriptor: | 1-NAPHTHOL, DNA REPAIR AND RECOMBINATION PROTEIN RADA, PHOSPHATE ION | | Authors: | Scott, D.E, Ehebauer, M.T, Pukala, T, Marsh, M, Blundell, T.L, Venkitaraman, A.R, Abell, C, Hyvonen, M. | | Deposit date: | 2012-07-20 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Using a Fragment-Based Approach to Target Protein-Protein Interactions.
Chembiochem, 14, 2013
|
|
4B34
 
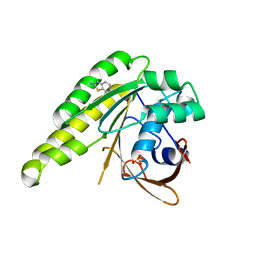 | | Humanised monomeric RadA in complex with 2-amino benzothiazole | | Descriptor: | 1,3-benzothiazol-2-amine, DNA REPAIR AND RECOMBINATION PROTEIN RADA, PHOSPHATE ION | | Authors: | Scott, D.E, Ehebauer, M.T, Pukala, T, Marsh, M, Blundell, T.L, Venkitaraman, A.R, Abell, C, Hyvonen, M. | | Deposit date: | 2012-07-20 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Using a Fragment-Based Approach to Target Protein-Protein Interactions.
Chembiochem, 14, 2013
|
|
4B3D
 
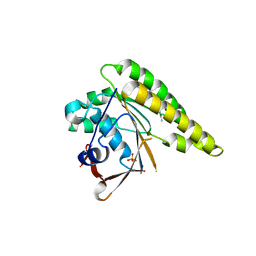 | | Humanised monomeric RadA in complex with 5-methyl indole | | Descriptor: | 5-METHYL INDOLE, DNA REPAIR AND RECOMBINATION PROTEIN RADA, PHOSPHATE ION | | Authors: | Scott, D.E, Ehebauer, M.T, Pukala, T, Marsh, M, Blundell, T.L, Venkitaraman, A.R, Abell, C, Hyvonen, M. | | Deposit date: | 2012-07-23 | | Release date: | 2013-02-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Using a Fragment-Based Approach to Target Protein-Protein Interactions.
Chembiochem, 14, 2013
|
|
1WFQ
 
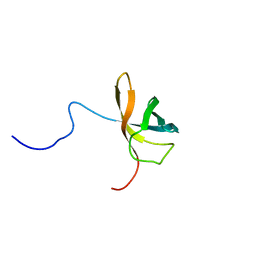 | | Solution structure of the first cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | UNR protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
2GD1
 
 | |
7L7Q
 
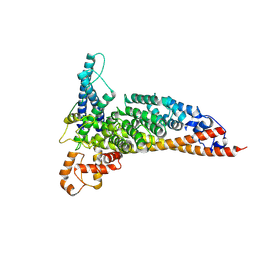 | | Ctf3c with Ulp2-KIM | | Descriptor: | Inner kinetochore subunit CTF3, Inner kinetochore subunit MCM16, Inner kinetochore subunit MCM22 | | Authors: | Hinshaw, S.M, Harrison, S.C. | | Deposit date: | 2020-12-30 | | Release date: | 2021-02-10 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Ctf3/CENP-I provides a docking site for the desumoylase Ulp2 at the kinetochore.
J.Cell Biol., 220, 2021
|
|
3ICE
 
 | | Rho transcription termination factor bound to RNA and ADP-BeF3 | | Descriptor: | 5'-R(P*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3', ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, ... | | Authors: | Thomsen, N.D, Berger, J.M. | | Deposit date: | 2009-07-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Running in reverse: the structural basis for translocation polarity in hexameric helicases.
Cell(Cambridge,Mass.), 139, 2009
|
|
2N1D
 
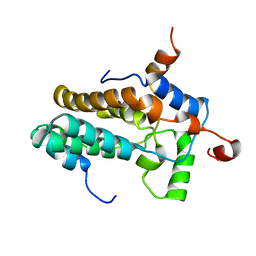 | | Solution structure of the MRG15-MRGBP complex | | Descriptor: | MRG/MORF4L-binding protein, Mortality factor 4-like protein 1 | | Authors: | Xie, T, Zmysloski, A.M, Zhang, Y, Radhakrishnan, I. | | Deposit date: | 2015-03-27 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Multi-specificity of MRG Domains.
Structure, 23, 2015
|
|
8TVU
 
 | |
3A5V
 
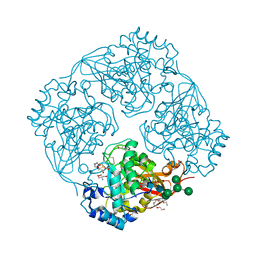 | | Crystal structure of alpha-galactosidase I from Mortierella vinacea | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fujimoto, Z, Kaneko, S, Kobayashi, H. | | Deposit date: | 2009-08-12 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Tetramer Structure of the Glycoside Hydrolase Family 27 alpha-Galactosidase I from Umbelopsis vinacea
Biosci.Biotechnol.Biochem., 73, 2009
|
|
3FPN
 
 | |
3JZG
 
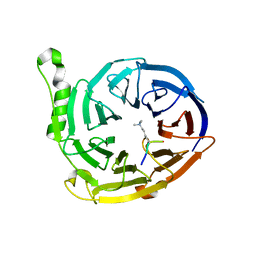 | | Structure of EED in complex with H3K27me3 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Xu, C, Bian, C.B, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
8U4U
 
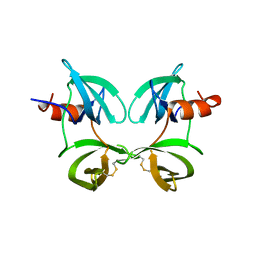 | |
3JZH
 
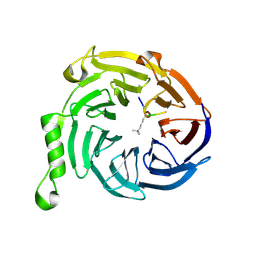 | | EED-H3K79me3 | | Descriptor: | HISTONE PEPTIDE, Polycomb protein EED | | Authors: | Xu, C, Bian, C.B, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-23 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Binding of different histone marks differentially regulates the activity and specificity of polycomb repressive complex 2 (PRC2).
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3DEE
 
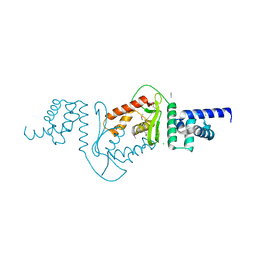 | |
3DGP
 
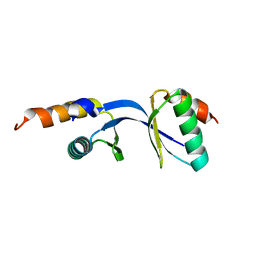 | | Crystal Structure of the complex between Tfb5 and the C-terminal domain of Tfb2 | | Descriptor: | RNA polymerase II transcription factor B subunit 2, RNA polymerase II transcription factor B subunit 5 | | Authors: | Kainov, D.E, Cavarelli, J, Egly, J.M, Poterszman, A. | | Deposit date: | 2008-06-14 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for group A trichothiodystrophy
Nat.Struct.Mol.Biol., 15, 2008
|
|
6FL4
 
 | | A. thaliana NUDT1 in complex with 8-oxo-dGTP | | Descriptor: | 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Nudix hydrolase 1 | | Authors: | Jemth, A.S, Scaletti-Hutchinson, E, Carter, M, Helleday, T, Stenmark, P. | | Deposit date: | 2018-01-25 | | Release date: | 2019-02-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure and Substrate Specificity of the 8-oxo-dGTP Hydrolase NUDT1 from Arabidopsis thaliana.
Biochemistry, 58, 2019
|
|
7XYF
 
 | |
