7NK3
 
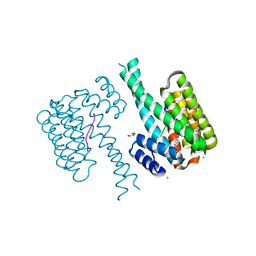 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-128 | | Descriptor: | 14-3-3 protein sigma, 2-(4-methylphenyl)sulfonyl-3,4-dihydro-1~{H}-isoquinoline, CALCIUM ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-17 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
8PPO
 
 | |
7NJ9
 
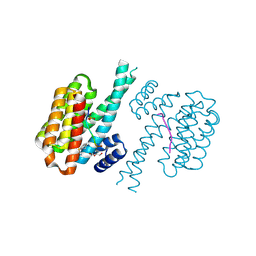 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-133 | | Descriptor: | 14-3-3 protein sigma, 4-(3,4-dihydro-2~{H}-quinolin-1-ylsulfonyl)benzaldehyde, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
7NIF
 
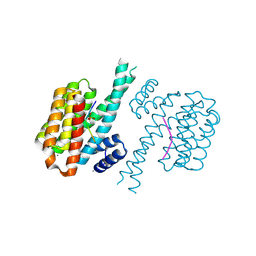 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound TCF521-011 | | Descriptor: | 1-(4-methylphenyl)imidazole, 14-3-3 protein sigma, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
4SDH
 
 | |
7NJB
 
 | | 14-3-3 sigma with RelA/p65 binding site pS45 and covalently bound TCF521-132 | | Descriptor: | 14-3-3 protein sigma, 4-piperidin-1-ylsulfonylbenzaldehyde, CHLORIDE ION, ... | | Authors: | Wolter, M, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | An Exploration of Chemical Properties Required for Cooperative Stabilization of the 14-3-3 Interaction with NF-kappa B-Utilizing a Reversible Covalent Tethering Approach.
J.Med.Chem., 64, 2021
|
|
1YCK
 
 | |
7NJ6
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1005 | | Descriptor: | 14-3-3 protein sigma, 4-[4-(trifluoromethyl)imidazol-1-yl]benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7NJ8
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1007 | | Descriptor: | 14-3-3 protein sigma, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
5DVA
 
 | |
7NIG
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1008 | | Descriptor: | 14-3-3 protein sigma, 2-bromanyl-4-imidazol-1-yl-benzaldehyde, CALCIUM ION, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-12 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
7NJA
 
 | | 14-3-3 sigma with Pin1 binding site pS72 and covalently bound LvD1006 | | Descriptor: | 14-3-3 protein sigma, CHLORIDE ION, Peptidyl-prolyl cis-trans isomerase NIMA-interacting 1, ... | | Authors: | Wolter, M, Dijck, L.v, Cossar, P.J, Ottmann, C. | | Deposit date: | 2021-02-16 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Reversible Covalent Imine-Tethering for Selective Stabilization of 14-3-3 Hub Protein Interactions.
J.Am.Chem.Soc., 143, 2021
|
|
4RML
 
 | | Crystal structure of the Olfactomedin domain of latrophilin 3 in C2221 crystal form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Latrophilin-3, MAGNESIUM ION | | Authors: | Ranaivoson, F.M, Liu, Q, Martini, F, Bergami, F, Von daake, S, Li, S, Demeler, B, Hendrickson, W.A, Comoletti, D. | | Deposit date: | 2014-10-21 | | Release date: | 2015-08-19 | | Last modified: | 2015-10-07 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Structural and Mechanistic Insights into the Latrophilin3-FLRT3 Complex that Mediates Glutamatergic Synapse Development.
Structure, 23, 2015
|
|
5DYU
 
 | | Crystal Structure of BAZ2B bromodomain in complex with fragment F39 | | Descriptor: | 1H-imidazol-5-ylmethanol, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Lolli, G, Caflisch, A. | | Deposit date: | 2015-09-25 | | Release date: | 2016-03-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-Throughput Fragment Docking into the BAZ2B Bromodomain: Efficient in Silico Screening for X-Ray Crystallography.
Acs Chem.Biol., 11, 2016
|
|
5DZ6
 
 | | Acyl transferase from Bacillaene PKS | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Till, M, Race, P.R. | | Deposit date: | 2015-09-25 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Architectural hierarchy of trans-acting enoyl reductases from polyunsaturated fatty acid and trans-acyltransferase polyketide synthases
To Be Published
|
|
4TLJ
 
 | | Ultra-high resolution crystal structure of caprine Beta-lactoglobulin | | Descriptor: | 1,4-BUTANEDIOL, Beta-lactoglobulin | | Authors: | Crowther, J.M, Jameson, G.B, Suzuki, H, Dobson, R.C.J. | | Deposit date: | 2014-05-30 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Ultra-high resolution crystal structure of recombinant caprine beta-lactoglobulin.
Febs Lett., 588, 2014
|
|
5E7B
 
 | | Structure of a nanobody (vHH) from camel against phage Tuc2009 RBP (BppL, ORF53) | | Descriptor: | nanobody nano-L06 | | Authors: | Legrand, P, Collins, B, Blangy, S, Murphy, J, Spinelli, S, Gutierrez, C, Richet, N, Kellenberger, C, Desmyter, A, Mahony, J, van Sinderen, D, Cambillau, C. | | Deposit date: | 2015-10-12 | | Release date: | 2015-12-30 | | Last modified: | 2016-05-04 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The Atomic Structure of the Phage Tuc2009 Baseplate Tripod Suggests that Host Recognition Involves Two Different Carbohydrate Binding Modules.
Mbio, 7, 2016
|
|
4HAX
 
 | |
4HB0
 
 | | Crystal structure of CRM1 inhibitor Leptomycin B in complex with CRM1(K541Q,K542Q,R543S,K545Q,K548Q,K579Q)-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Sun, Q, Chook, Y.M. | | Deposit date: | 2012-09-27 | | Release date: | 2013-01-09 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nuclear export inhibition through covalent conjugation and hydrolysis of Leptomycin B by CRM1.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
6RWH
 
 | | Fragment AZ-007 binding at a primary and secondary binding site of the the p53pT387/14-3-3 sigma complex | | Descriptor: | 14-3-3 protein sigma, 5-(1~{H}-imidazol-5-yl)-4-phenyl-thiophene-2-carboximidamide, CALCIUM ION, ... | | Authors: | Leysen, S, Guillory, X, Wolter, M, Genet, S, Somsen, B, Patel, J, Castaldi, P, Ottmann, C. | | Deposit date: | 2019-06-05 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Fragment-based Differential Targeting of PPI Stabilizer Interfaces.
J.Med.Chem., 63, 2020
|
|
1HIM
 
 | |
7LPA
 
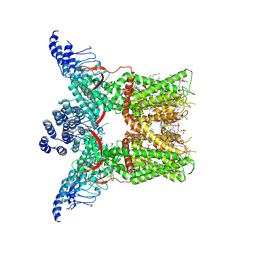 | | Cryo-EM structure of full-length TRPV1 with capsaicin at 4 degrees Celsius | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7LPE
 
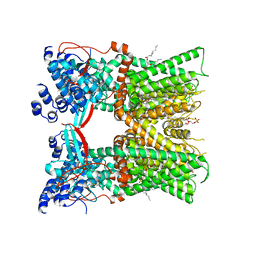 | | Cryo-EM structure of full-length TRPV1 with capsaicin at 48 degrees Celsius, in an open state, class 1 | | Descriptor: | (6E)-N-(4-hydroxy-3-methoxybenzyl)-8-methylnon-6-enamide, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, Transient receptor potential cation channel subfamily V member 1, ... | | Authors: | Kwon, D.H, Zhang, F, Suo, Y, Lee, S.-Y. | | Deposit date: | 2021-02-11 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Heat-dependent opening of TRPV1 in the presence of capsaicin.
Nat.Struct.Mol.Biol., 28, 2021
|
|
1HIN
 
 | |
4QRP
 
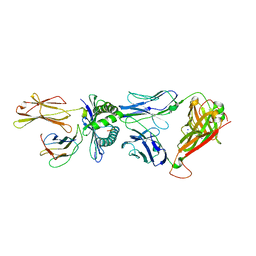 | | Crystal Structure of HLA B*0801 in complex with HSKKKCDEL and DD31 TCR | | Descriptor: | Beta-2-microglobulin, DD31 TCR alpha chain, DD31 TCR beta chain, ... | | Authors: | Gras, S, Berry, R, Lucet, I.S, Rossjohn, J. | | Deposit date: | 2014-07-02 | | Release date: | 2014-11-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | An Extensive Antigenic Footprint Underpins Immunodominant TCR Adaptability against a Hypervariable Viral Determinant.
J.Immunol., 193, 2014
|
|
