1I70
 
 | | CRYSTAL STRUCTURE OF RNASE SA Y86F MUTANT | | Descriptor: | GUANYL-SPECIFIC RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Urbanikova, L. | | Deposit date: | 2001-03-07 | | Release date: | 2001-09-19 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Tyrosine hydrogen bonds make a large contribution to protein stability.
J.Mol.Biol., 312, 2001
|
|
6YUN
 
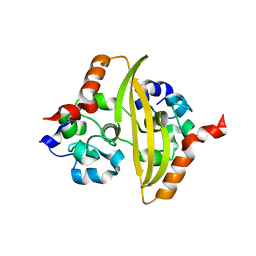 | |
6ZCO
 
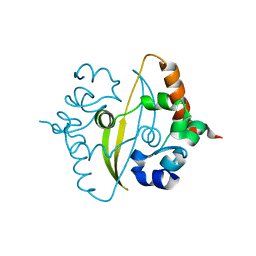 | | Crystal Structure of C-terminal Dimerization Domain of Nucleocapsid Phosphoprotein from SARS-CoV-2, crystal form II | | Descriptor: | Nucleoprotein | | Authors: | Zinzula, L, Basquin, J, Nagy, I, Bracher, A. | | Deposit date: | 2020-06-11 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.361 Å) | | Cite: | High-resolution structure and biophysical characterization of the nucleocapsid phosphoprotein dimerization domain from the Covid-19 severe acute respiratory syndrome coronavirus 2.
Biochem.Biophys.Res.Commun., 538, 2021
|
|
3BIW
 
 | | Crystal structure of the Neuroligin-1/Neurexin-1beta synaptic adhesion complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Arac, D, Boucard, A.A, Ozkan, E, Strop, P, Newell, E, Sudhof, T.C, Brunger, A.T. | | Deposit date: | 2007-12-01 | | Release date: | 2007-12-18 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structures of Neuroligin-1 and the Neuroligin-1/Neurexin-1beta Complex Reveal Specific Protein-Protein and Protein-Ca(2+) Interactions.
Neuron, 56, 2007
|
|
1JXB
 
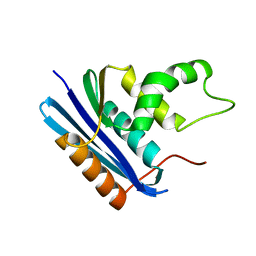 | |
1LAJ
 
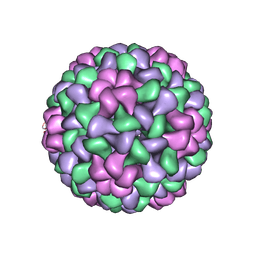 | | The Structure of Tomato Aspermy Virus by X-Ray Crystallography | | Descriptor: | 5'-R(*AP*AP*A)-3', MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Lucas, R.W, Larson, S.B, Canady, M.A, McPherson, A. | | Deposit date: | 2002-03-28 | | Release date: | 2002-11-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The Structure of Tomato Aspermy Virus by X-Ray Crystallography
J.STRUCT.BIOL., 139, 2002
|
|
3GCA
 
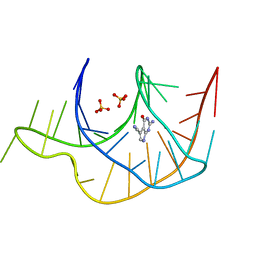 | |
2XWH
 
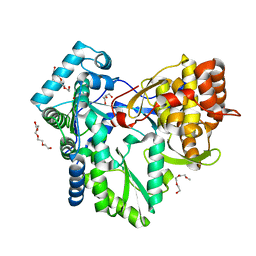 | | HCV-J6 NS5B polymerase structure at 1.8 Angstrom | | Descriptor: | DI(HYDROXYETHYL)ETHER, POLYETHYLENE GLYCOL (N=34), RNA DEPENDENT RNA POLYMERASE | | Authors: | Scrima, N, Bressanelli, S. | | Deposit date: | 2010-11-03 | | Release date: | 2011-01-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Comprehensive Structure-Function Comparison of Hepatitis C Virus Strains Jfh1 and J6 Polymerases Reveals a Key Residue Stimulating Replication in Cell Culture Across Genotypes.
J.Virol., 85, 2011
|
|
5V37
 
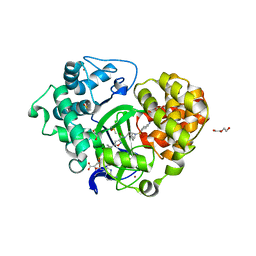 | | Crystal structure of SMYD3 with SAM and EPZ028862 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Boriack-Sjodin, P.A. | | Deposit date: | 2017-03-06 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Small molecule inhibitors and CRISPR/Cas9 mutagenesis demonstrate that SMYD2 and SMYD3 activity are dispensable for autonomous cancer cell proliferation.
Plos One, 13, 2018
|
|
2Z41
 
 | | Crystal Structure Analysis of the Ski2-type RNA helicase | | Descriptor: | MAGNESIUM ION, putative ski2-type helicase | | Authors: | Nakashima, T, Zhang, X, Kakuta, Y, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2007-06-12 | | Release date: | 2008-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.51 Å) | | Cite: | Crystal structure of an archaeal Ski2p-like protein from Pyrococcus horikoshii OT3
Protein Sci., 17, 2008
|
|
5VI7
 
 | |
2GJK
 
 | | Structural and functional insights into the human Upf1 helicase core | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Regulator of nonsense transcripts 1 | | Authors: | Cheng, Z, Muhlrad, D, Parker, R, Song, H. | | Deposit date: | 2006-03-31 | | Release date: | 2007-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and functional insights into the human Upf1 helicase core
Embo J., 26, 2007
|
|
2GK6
 
 | | Structural and Functional insights into the human Upf1 helicase core | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Cheng, Z, Muhlrad, D, Parker, R, Song, H. | | Deposit date: | 2006-03-31 | | Release date: | 2007-01-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and functional insights into the human Upf1 helicase core
Embo J., 26, 2007
|
|
3UCU
 
 | | The c-di-GMP-I riboswitch bound to pGpG | | Descriptor: | MAGNESIUM ION, RNA (92-MER), U1 small nuclear ribonucleoprotein A, ... | | Authors: | Smith, K.D, Strobel, S.A. | | Deposit date: | 2011-10-27 | | Release date: | 2012-01-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical characterization of linear dinucleotide analogues bound to the c-di-GMP-I aptamer.
Biochemistry, 51, 2012
|
|
2QJ1
 
 | | Crystal structure of infectious bursal disease virus VP1 polymerase incubated with an oligopeptide mimicking the VP3 C-terminus | | Descriptor: | Infectious bursal disease virus VP1 polymerase | | Authors: | Garriga, D, Navarro, A, Querol-Audi, J, Abaitua, F, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2007-07-06 | | Release date: | 2007-11-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.48 Å) | | Cite: | Unprecedented activation mechanism of a non-canonical RNA-dependent RNA polymerase
To be Published
|
|
3IQR
 
 | |
7V5Y
 
 | |
7V5Z
 
 | |
3IQN
 
 | |
3IQP
 
 | |
5XPG
 
 | | Crystal structure of T. thermophilus Argonaute protein complexed with a bulge 6'U7' on the target strand | | Descriptor: | 5'-D(P*TP*GP*AP*GP*GP*TP*AP*GP*TP*AP*GP*GP*TP*TP*GP*TP*AP*TP*A P*GP*T)-3', 5'-R(*UP*AP*U*AP*CP*AP*AP*CP*CP*UP*AP*CP*AP*UP*AP*CP*CP*UP*CP* G)-3', MAGNESIUM ION, ... | | Authors: | Sheng, G, Gogakos, T, Wang, J, Zhao, H, Serganov, A, Juranek, S, Tuschl, T, Patel, J.D, Wang, Y. | | Deposit date: | 2017-06-02 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes.
Nucleic Acids Res., 45, 2017
|
|
4HZR
 
 | | Crystal structure of Ack1 kinase domain | | Descriptor: | 1,2-ETHANEDIOL, Activated CDC42 kinase 1, CHLORIDE ION, ... | | Authors: | Gajiwala, K.S. | | Deposit date: | 2012-11-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Ack1: activation and regulation by allostery.
Plos One, 8, 2013
|
|
4I59
 
 | |
2X25
 
 | | Free acetyl-CypA orthorhombic form | | Descriptor: | PEPTIDYL-PROLYL CIS-TRANS ISOMERASE A | | Authors: | Lammers, M, Neumann, H, Chin, J.W, James, L.C. | | Deposit date: | 2010-01-11 | | Release date: | 2010-03-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Acetylation Regulates Cyclophilin a Catalysis, Immunosuppression and HIV Isomerization.
Nat.Chem.Biol., 6, 2010
|
|
4I58
 
 | |
