6PE3
 
 | |
6PKD
 
 | | Myocilin OLF mutant N428D/D478H | | Descriptor: | Myocilin, SODIUM ION | | Authors: | Lieberman, R.L, Hill, S.E. | | Deposit date: | 2019-06-29 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calcium-ligand variants of the myocilin olfactomedin propeller selected from invertebrate phyla reveal cross-talk with N-terminal blade and surface helices.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6V4R
 
 | |
6YPF
 
 | | NUDIX1 hydrolase from Rosa x hybrida in complex with geranyl pyrophosphate | | Descriptor: | GERANYL DIPHOSPHATE, Geranyl diphosphate phosphohydrolase | | Authors: | Degut, C, Rety, S, Tisne, C, Baudino, S. | | Deposit date: | 2020-04-16 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Functional diversification in the Nudix hydrolase gene family drives sesquiterpene biosynthesis in Rosa × wichurana.
Plant J., 104, 2020
|
|
5YFB
 
 | | Crystal structure of a new DPP III family member | | Descriptor: | 1,2-ETHANEDIOL, Dipeptidyl peptidase 3, GLYCEROL, ... | | Authors: | Xu, T, Sun, Z, Liu, J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Aflatoxin-oxidase from Armillariella tabescens reveal a dual activity enzyme.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
5Y62
 
 | | YfiR complexed with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, YfiR | | Authors: | Zhou, L, Xu, M, Jiang, T. | | Deposit date: | 2017-08-10 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into the functional role of GMP in modulating the YfiBNR system
Biochem. Biophys. Res. Commun., 493, 2017
|
|
6OM8
 
 | | Caenorhabditis Elegans UDP-Glucose Dehydrogenase in complex with UDP-Xylose | | Descriptor: | UDP-glucose 6-dehydrogenase, URIDINE-5'-DIPHOSPHATE-XYLOPYRANOSE | | Authors: | Beattie, N.R, McDonald, W.E, Hicks Sirmans, T.N, Wood, Z.A. | | Deposit date: | 2019-04-18 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.449 Å) | | Cite: | Conservation of Atypical Allostery inC. elegansUDP-Glucose Dehydrogenase.
Acs Omega, 4, 2019
|
|
6V4Q
 
 | | Crystal structure of a MR78-like antibody naive-1 Fab | | Descriptor: | GLYCEROL, ISOPROPYL ALCOHOL, Naive-1 Fab heavy chain, ... | | Authors: | Bozhanova, N.G, Crowe, J.E, Meiler, J. | | Deposit date: | 2019-11-28 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Discovery of Marburg virus neutralizing antibodies from virus-naive human antibody repertoires using large-scale structural predictions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5YHJ
 
 | | Cytochrome P450EX alpha (CYP152N1) wild-type with myristic acid | | Descriptor: | Cytochrome P450, MYRISTIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Onoda, H, Shoji, O, Suzuki, K, Sugimoto, H, Shiro, Y, Watanabe, Y. | | Deposit date: | 2017-09-28 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Alpha-Oxidative Decarboxylation of Fatty Acids Catalysed by Cytochrome P450 Peroxygenases Yielding Shorter-Alkyl-Chain Fatty Acids
Catalysis Science And Technology, 2017
|
|
6O67
 
 | |
5YGV
 
 | | Crystal structure of the abscisic acid receptor PYR1 in complex with an antagonist | | Descriptor: | (2Z,4E)-3-methyl-5-[(1S,4S)-2,6,6-trimethyl-4-[3-(4-methylphenyl)prop-2-ynoxy]-1-oxidanyl-cyclohex-2-en-1-yl]penta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Akiyama, T, Sue, M, Takeuchi, J, Mimura, N, Okamoto, M, Monda, K, Iba, K, Ohnishi, T, Todoroki, Y, Yajima, S. | | Deposit date: | 2017-09-27 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Chemical Design of Abscisic Acid Antagonists That Block PYL-PP2C Receptor Interactions.
ACS Chem. Biol., 13, 2018
|
|
6QA7
 
 | | Glycogen Phosphorylase b in complex with 29 | | Descriptor: | (5~{S},7~{R},8~{S},9~{S},10~{R})-7-(hydroxymethyl)-2-naphthalen-1-yl-8,9,10-tris(oxidanyl)-6-oxa-1,3-diazaspiro[4.5]dec-1-en-4-one, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Glucopyranosylidene-spiro-imidazolinones, a New Ring System: Synthesis and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetics and X-ray Crystallography.
J.Med.Chem., 62, 2019
|
|
6O5D
 
 | | PYOCHELIN | | Descriptor: | GLYCEROL, Neutrophil gelatinase-associated lipocalin, SULFATE ION | | Authors: | Rupert, P.B, Strong, R.K, Clifton, M.C, Edwards, T.E, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2019-03-01 | | Release date: | 2019-09-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Parsing the functional specificity of Siderocalin/Lipocalin 2/NGAL for siderophores and related small-molecule ligands.
J Struct Biol X, 2, 2019
|
|
5YFC
 
 | | Crystal structure of a new DPP III family member | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Dipeptidyl peptidase 3, ... | | Authors: | Xu, T, Liu, J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structures of Aflatoxin-oxidase from Armillariella tabescens reveal a dual activity enzyme.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
6O9U
 
 | | KirBac3.1 at a resolution of 2 Angstroms | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3,3',3''-phosphoryltripropanoic acid, BARIUM ION, ... | | Authors: | Gulbis, J.M, Clarke, O.B. | | Deposit date: | 2019-03-15 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A constricted opening in Kir channels does not impede potassium conduction.
Nat Commun, 11, 2020
|
|
5YFD
 
 | | Crystal structure of a new DPP III family member | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, COPPER (II) ION, Dipeptidyl peptidase 3, ... | | Authors: | Xu, T, Liu, J. | | Deposit date: | 2017-09-20 | | Release date: | 2018-01-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structures of Aflatoxin-oxidase from Armillariella tabescens reveal a dual activity enzyme.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
5Y61
 
 | | YfiB-YfiR complexed with GMP | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, YfiB, YfiR | | Authors: | Zhou, L, Xu, M, Jiang, T. | | Deposit date: | 2017-08-10 | | Release date: | 2017-09-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural insights into the functional role of GMP in modulating the YfiBNR system
Biochem. Biophys. Res. Commun., 493, 2017
|
|
6QA6
 
 | | Glycogen Phosphorylase b in complex with 30 | | Descriptor: | (5~{S},7~{R},8~{S},9~{S},10~{R})-7-(hydroxymethyl)-2-naphthalen-2-yl-8,9,10-tris(oxidanyl)-6-oxa-1,3-diazaspiro[4.5]dec-1-en-4-one, DIMETHYL SULFOXIDE, Glycogen phosphorylase, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Glucopyranosylidene-spiro-imidazolinones, a New Ring System: Synthesis and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetics and X-ray Crystallography.
J.Med.Chem., 62, 2019
|
|
6XU4
 
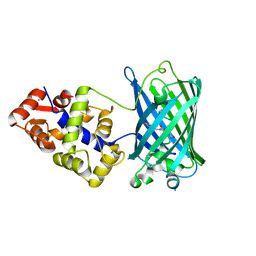 | | Crystal structure of the genetically-encoded FGCaMP calcium indicator in its calcium-bound state | | Descriptor: | CALCIUM ION, FGCamp | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Korzhenevskiy, D.A, Barykina, N.V, Subach, O.M, Subach, F.V. | | Deposit date: | 2020-01-17 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | FGCaMP7, an Improved Version of Fungi-Based Ratiometric Calcium Indicator for In Vivo Visualization of Neuronal Activity.
Int J Mol Sci, 21, 2020
|
|
6O68
 
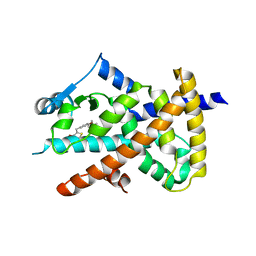 | |
6O6C
 
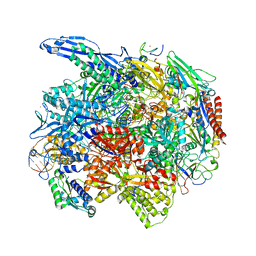 | | RNA polymerase II elongation complex arrested at a CPD lesion | | Descriptor: | DNA (27-MER), DNA (5'-D(P*GP*GP*AP*GP*AP*AP*GP*GP*AP*GP*CP*AP*GP*AP*GP*C)-3'), DNA-directed RNA polymerase II subunit RPB1, ... | | Authors: | Lahiri, I, Leshziner, A.E. | | Deposit date: | 2019-03-05 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | 3.1 angstrom structure of yeast RNA polymerase II elongation complex stalled at a cyclobutane pyrimidine dimer lesion solved using streptavidin affinity grids.
J.Struct.Biol., 207, 2019
|
|
6QA8
 
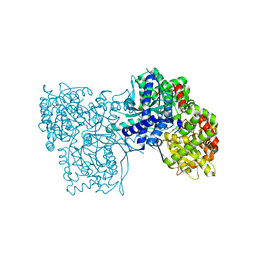 | | Glycogen Phosphorylase b in complex with 28 | | Descriptor: | (5~{S},7~{R},8~{S},9~{S},10~{R})-7-(hydroxymethyl)-8,9,10-tris(oxidanyl)-2-phenyl-6-oxa-1,3-diazaspiro[4.5]dec-1-en-4-one, Glycogen phosphorylase, muscle form, ... | | Authors: | Kyriakis, E, Stravodimos, G.A, Skamnaki, V.T, Leonidas, D.D. | | Deposit date: | 2018-12-18 | | Release date: | 2019-06-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Glucopyranosylidene-spiro-imidazolinones, a New Ring System: Synthesis and Evaluation as Glycogen Phosphorylase Inhibitors by Enzyme Kinetics and X-ray Crystallography.
J.Med.Chem., 62, 2019
|
|
5YIO
 
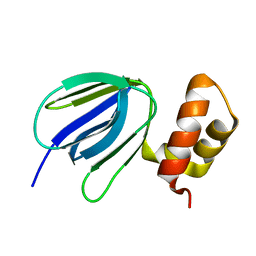 | | NMR solution structure of subunit epsilon of the Mycobacterium tuberculosis F-ATP synthase | | Descriptor: | ATP synthase epsilon chain | | Authors: | Shin, J, Ragunathan, P, Sundararaman, L, Nartey, W, Manimekalai, M.S.S, Bogdanovic, N, Gruber, G. | | Deposit date: | 2017-10-06 | | Release date: | 2018-10-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structure of Mycobacterium tuberculosis F-ATP synthase subunit epsilon provides new insight into energy coupling inside the rotary engine.
FEBS J., 285, 2018
|
|
6QE6
 
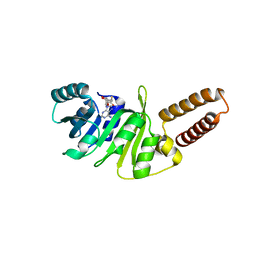 | | Structure of M. capricolum TrmK in complex with the natural cofactor product S-adenosyl-homocysteine (SAH) | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, tRNA (Adenine(22)-N(1))-methyltransferase | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
6QE5
 
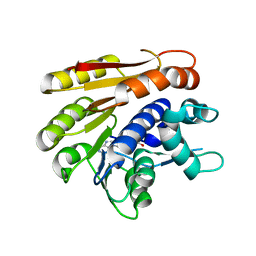 | | Structure of E.coli RlmJ in complex with the natural cofactor product S-adenosyl-homocysteine (SAH) | | Descriptor: | Ribosomal RNA large subunit methyltransferase J, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Oerum, S, Catala, M, Atdjian, C, Brachet, F, Ponchon, L, Barraud, P, Iannazzo, L, Droogmans, L, Braud, E, Etheve-Quelquejeu, M, Tisne, C. | | Deposit date: | 2019-01-04 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Bisubstrate analogues as structural tools to investigate m6A methyltransferase active sites.
Rna Biol., 16, 2019
|
|
