4NLU
 
 | | Poliovirus Polymerase - G289A Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4JLQ
 
 | | Crystal structure of human Karyopherin-beta2 bound to the PY-NLS of Saccharomyces cerevisiae NAB2 | | Descriptor: | Nuclear polyadenylated RNA-binding protein NAB2, Transportin-1 | | Authors: | Sampathkumar, P, Gizzi, A, Rout, M.P, Chook, Y.M, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Nucleocytoplasmic Transport: a Target for Cellular Control (NPCXstals) | | Deposit date: | 2013-03-12 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structure of human Karyopherin beta 2 bound to the PY-NLS of Saccharomyces cerevisiae Nab2.
J.Struct.Funct.Genom., 14, 2013
|
|
4NLP
 
 | | Poliovirus Polymerase - C290V Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
1P9X
 
 | | THE CRYSTAL STRUCTURE OF THE 50S LARGE RIBOSOMAL SUBUNIT FROM DEINOCOCCUS RADIODURANS COMPLEXED WITH TELITHROMYCIN KETOLIDE ANTIBIOTIC | | Descriptor: | 23S RIBOSOMAL RNA, TELITHROMYCIN | | Authors: | Berisio, R, Harms, J, Schluenzen, F, Zarivach, R, Hansen, H.A, Fucini, P, Yonath, A. | | Deposit date: | 2003-05-13 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural insight into the antibiotic action of telithromycin against resistant mutants
J.Bacteriol., 185, 2003
|
|
1A8V
 
 | |
5V1M
 
 | | Structure of human Usb1 with uridine 5'-monophosphate | | Descriptor: | CHLORIDE ION, GLYCEROL, U6 snRNA phosphodiesterase, ... | | Authors: | Montemayor, E.J, Didychuk, A.L, Butcher, S.E. | | Deposit date: | 2017-03-02 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Usb1 controls U6 snRNP assembly through evolutionarily divergent cyclic phosphodiesterase activities.
Nat Commun, 8, 2017
|
|
4NLO
 
 | | Poliovirus Polymerase - C290I Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NYH
 
 | | Orthorhombic crystal form of pir1 dual specificity phosphatase core | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, RNA/RNP complex-1-interacting phosphatase | | Authors: | Sankhala, R.S, Lokareddy, R.K, Cingolani, G. | | Deposit date: | 2013-12-10 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Human PIR1, an Atypical Dual-Specificity Phosphatase.
Biochemistry, 53, 2014
|
|
8U7B
 
 | |
2A6T
 
 | |
1DFJ
 
 | |
3ZD4
 
 | |
4NLW
 
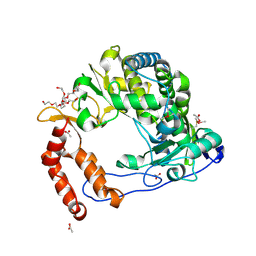 | | Poliovirus Polymerase - G289A/C290I Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NLY
 
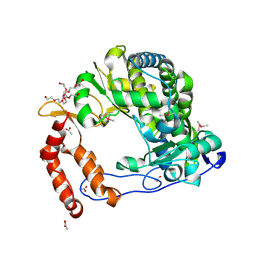 | | Poliovirus Polymerase - C290E Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
4NLR
 
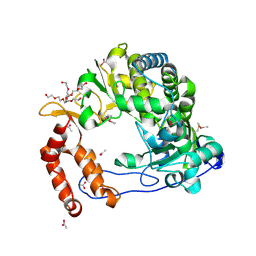 | | Poliovirus Polymerase - C290S Loop Mutant | | Descriptor: | ACETIC ACID, PENTAETHYLENE GLYCOL, RNA-directed RNA polymerase 3D-POL, ... | | Authors: | Sholders, A.J, Peersen, O.B. | | Deposit date: | 2013-11-14 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Distinct conformations of a putative translocation element in poliovirus polymerase.
J.Mol.Biol., 426, 2014
|
|
3V2K
 
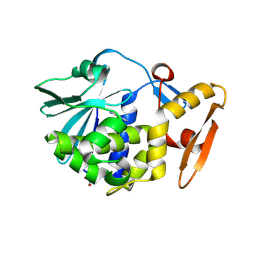 | | Crystal structure of ribosome inactivating protein from momordica balsamina complexed with the product of RNA substrate adenosine triphosphate at 2.0 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENINE, GLYCEROL, ... | | Authors: | Kushwaha, G.S, Pandey, N, Sinha, M, Bhushan, A, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2011-12-12 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structures of a type-1 ribosome inactivating protein from Momordica balsamina in the bound and unbound states.
Biochim.Biophys.Acta, 1824, 2012
|
|
4L81
 
 | | Structure of the SAM-I/IV riboswitch (env87(deltaU92, deltaG93)) | | Descriptor: | COBALT HEXAMMINE(III), MAGNESIUM ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Trausch, J.J, Reyes, F.E, Edwards, A.L, Batey, R.T. | | Deposit date: | 2013-06-15 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural basis for diversity in the SAM clan of riboswitches.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7ZIP
 
 | |
7ZIQ
 
 | | BK Polyomavirus VP1 in complex with 6'-Sialyllactose glycomacromolecules (aromatic linker) | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein VP1, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freytag, J, Mueller, J.C, Stehle, T. | | Deposit date: | 2022-04-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis of Homo- and Heteromultivalent Fucosylated and Sialylated Oligosaccharide Conjugates via Preactivated N -Methyloxyamine Precision Macromolecules and Their Binding to Polyomavirus Capsid Proteins.
Biomacromolecules, 23, 2022
|
|
7ZIN
 
 | | JC Polyomavirus VP1 in complex with 6'-Sialyllactose glycomacromolecules (aliphatic linker) | | Descriptor: | 1,2-ETHANEDIOL, Major capsid protein VP1, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose, ... | | Authors: | Freytag, J, Mueller, J.C, Stehle, T. | | Deposit date: | 2022-04-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Synthesis of Homo- and Heteromultivalent Fucosylated and Sialylated Oligosaccharide Conjugates via Preactivated N -Methyloxyamine Precision Macromolecules and Their Binding to Polyomavirus Capsid Proteins.
Biomacromolecules, 23, 2022
|
|
7ZIM
 
 | | JC Polyomavirus VP1 in complex with 3'-Sialyllactose glycomacromolecules (aromatic linker) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Major capsid protein VP1, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose, ... | | Authors: | Freytag, J, Mueller, J.C, Stehle, T. | | Deposit date: | 2022-04-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Synthesis of Homo- and Heteromultivalent Fucosylated and Sialylated Oligosaccharide Conjugates via Preactivated N -Methyloxyamine Precision Macromolecules and Their Binding to Polyomavirus Capsid Proteins.
Biomacromolecules, 23, 2022
|
|
7ZIO
 
 | | JC Polyomavirus VP1 in complex with 6'-Sialyllactose glycomacromolecules (aromatic linker) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freytag, J, Mueller, J.C, Stehle, T. | | Deposit date: | 2022-04-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Synthesis of Homo- and Heteromultivalent Fucosylated and Sialylated Oligosaccharide Conjugates via Preactivated N -Methyloxyamine Precision Macromolecules and Their Binding to Polyomavirus Capsid Proteins.
Biomacromolecules, 23, 2022
|
|
7ZIL
 
 | | JC Polyomavirus VP1 in complex with 3'-Sialyllactose glycomacromolecules (aliphatic linker) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Freytag, J, Mueller, J.C, Stehle, T. | | Deposit date: | 2022-04-08 | | Release date: | 2022-12-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Synthesis of Homo- and Heteromultivalent Fucosylated and Sialylated Oligosaccharide Conjugates via Preactivated N -Methyloxyamine Precision Macromolecules and Their Binding to Polyomavirus Capsid Proteins.
Biomacromolecules, 23, 2022
|
|
1F3L
 
 | |
5CFD
 
 | |
