3LVX
 
 | |
3LY9
 
 | |
3LY7
 
 | |
6ICD
 
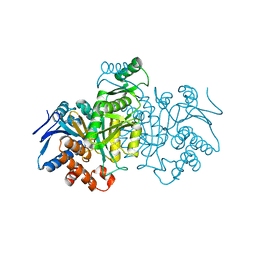 | | REGULATION OF AN ENZYME BY PHOSPHORYLATION AT THE ACTIVE SITE | | Descriptor: | ISOCITRATE DEHYDROGENASE | | Authors: | Hurley, J.H, Dean, A.M, Sohl, J.L, Koshlandjunior, D.E, Stroud, R.M. | | Deposit date: | 1990-05-30 | | Release date: | 1991-10-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Regulation of an enzyme by phosphorylation at the active site.
Science, 249, 1990
|
|
8CY8
 
 | |
8DR4
 
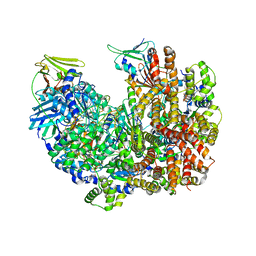 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) without NTD | | Descriptor: | DNA (5'-D(P*AP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.45 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR3
 
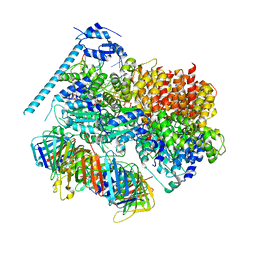 | | Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR5
 
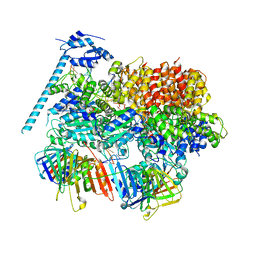 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) with NTD | | Descriptor: | DNA (5'-D(P*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*G)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR1
 
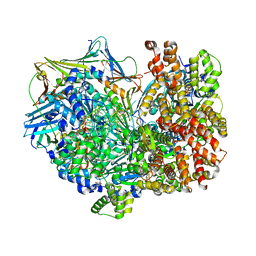 | | Consensus closed state of RFC:PCNA bound to a 3' ss/dsDNA junction (DNA2) | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*CP*CP*CP*CP*CP*CP*TP*TP*T)-3'), DNA (5'-D(P*CP*CP*CP*CP*CP*CP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*AP*GP*GP*GP*GP*GP*GP*GP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-17 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.14 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DQW
 
 | |
8SWM
 
 | |
7ANG
 
 | |
7ANC
 
 | |
8DQZ
 
 | | Intermediate state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DR0
 
 | | Closed state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(P*CP*CP*CP*CP*GP*GP*GP*GP*CP*CP*CP*CP*CP*CP*CP*GP*GP*C)-3'), DNA (5'-D(P*TP*TP*TP*TP*TP*TP*CP*GP*GP*GP*GP*GP*GP*GP*CP*CP*CP*CP*GP*GP*GP*G)-3'), GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
8DQX
 
 | | Open state of RFC:PCNA bound to a 3' ss/dsDNA junction | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*T)-3'), DNA (5'-D(P*TP*CP*CP*GP*AP*GP*CP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*TP*GP*CP*CP*CP*GP*GP*A)-3'), ... | | Authors: | Schrecker, M, Hite, R.K. | | Deposit date: | 2022-07-20 | | Release date: | 2022-08-24 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Multistep loading of a DNA sliding clamp onto DNA by replication factor C.
Elife, 11, 2022
|
|
2WV5
 
 | | Crystal structure of foot-and-mouth disease virus 3C protease in complex with a decameric peptide corresponding to the VP1-2A cleavage junction with a GLN to Glu substitution at P1 | | Descriptor: | FOOT AND MOUTH DISEASE VIRUS (SEROTYPE A) VARIANT VP1 CAPSID PROTEIN, PICORNAIN 3C | | Authors: | Zunszain, P.A, Knox, S.R, Sweeney, T.R, Yang, J, Roque-Rosell, N, Belsham, G.J, Leatherbarrow, R.J, Curry, S. | | Deposit date: | 2009-10-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights Into Cleavage Specificity from the Crystal Structure of Foot-and-Mouth Disease Virus 3C Protease Complexed with a Peptide Substrate.
J.Mol.Biol., 395, 2010
|
|
2WV4
 
 | | Crystal structure of foot-and-mouth disease virus 3C protease in complex with a decameric peptide corresponding to the VP1-2A cleavage junction | | Descriptor: | FOOT AND MOUTH DISEASE VIRUS (SEROTYPE A) VARIANT VP1 CAPSID PROTEIN, PICORNAIN 3C | | Authors: | Zunszain, P.A, Knox, S.R, Sweeney, T.R, Yang, J, Roque-Rosell, N, Belsham, G.J, Leatherbarrow, R.J, Curry, S. | | Deposit date: | 2009-10-13 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into Cleavage Specificity from the Crystal Structure of Foot-and-Mouth Disease Virus 3C Protease Complexed with a Peptide Substrate.
J.Mol.Biol., 395, 2010
|
|
7AN4
 
 | |
7ANJ
 
 | |
8TP9
 
 | | H2 hemagglutinin (A/Singapore/1/1957) in complex with medial-junction-targeting Fab 2-2-1G06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 2-2-1G06, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
8TPA
 
 | | H1 hemagglutinin (NC99) in complex with medial-junction-targeting Fab 2-2-1G06 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of Fab 2-2-1G06, ... | | Authors: | Yang, Y.R, Han, J, Perrett, H.R, Ward, A.B. | | Deposit date: | 2023-08-04 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Immune memory shapes human polyclonal antibody responses to H2N2 vaccination.
Biorxiv, 2023
|
|
7ANI
 
 | |
2VCG
 
 | | Crystal structure of a HDAC-like protein HDAH from Bordetella sp. with the bound inhibitor ST-17 | | Descriptor: | CHLORIDE ION, GLYCEROL, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Dickmanns, A, Strasser, A, Ficner, R. | | Deposit date: | 2007-09-24 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phenylalanine-Containing Hydroxamic Acids as Selective Inhibitors of Class Iib Histone Deacetylases (Hdacs).
Bioorg.Med.Chem., 16, 2008
|
|
6HBI
 
 | | SCAPHARCA DIMERIC HEMOGLOBIN, MUTANT T72V, DEOXY FORM | | Descriptor: | HEMOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Royer Junior, W.E. | | Deposit date: | 1998-06-25 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mutational destabilization of the critical interface water cluster in Scapharca dimeric hemoglobin: structural basis for altered allosteric activity.
J.Mol.Biol., 284, 1998
|
|
