4C0D
 
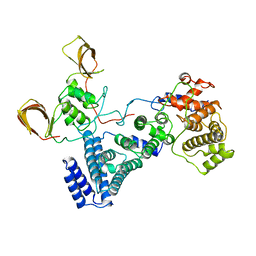 | | Structure of the NOT module of the human CCR4-NOT complex (CNOT1-CNOT2-CNOT3) | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 2, CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 3 | | Authors: | Raisch, T, Jonas, S, Boland, A, Chen, Y, Izaurralde, E, Weichenrieder, O. | | Deposit date: | 2013-08-01 | | Release date: | 2013-10-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and Assembly of the not Module of the Human Ccr4-not Complex
Nat.Struct.Mol.Biol., 20, 2013
|
|
8BFI
 
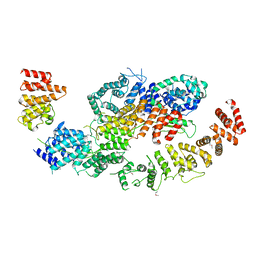 | | human CNOT1-CNOT10-CNOT11 module | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 10, CCR4-NOT transcription complex subunit 11 | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2022-10-26 | | Release date: | 2022-12-28 | | Last modified: | 2023-02-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The human CNOT1-CNOT10-CNOT11 complex forms a structural platform for protein-protein interactions.
Cell Rep, 42, 2023
|
|
4GMJ
 
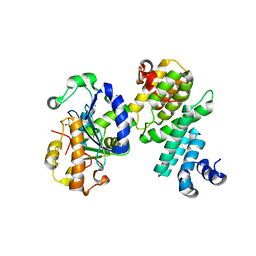 | | Structure of human NOT1 MIF4G domain co-crystallized with CAF1 | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 7, CHLORIDE ION, ... | | Authors: | Petit, P, Weichenrieder, O, Wohlbold, L, Izaurralde, E. | | Deposit date: | 2012-08-16 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex
Nucleic Acids Res., 40, 2012
|
|
4CT7
 
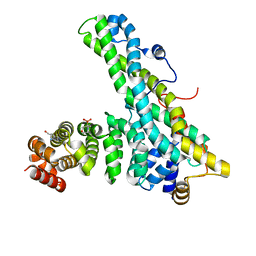 | | CNOT9-CNOT1 complex with bound tryptophan | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, CELL DIFFERENTIATION PROTEIN RCD1 HOMOLOG, ... | | Authors: | Basquin, J, Ozgur, S, Conti, E. | | Deposit date: | 2014-03-12 | | Release date: | 2014-05-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.897 Å) | | Cite: | Structural and Biochemical Insights to the Role of the Ccr4-not Complex and Ddx6 ATPase in Microrna Repression.
Mol.Cell, 54, 2014
|
|
5ANR
 
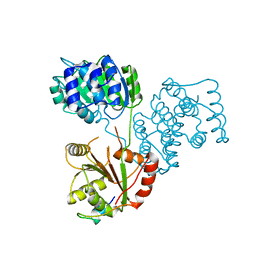 | | Structure of a human 4E-T - DDX6 - CNOT1 complex | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E TRANSPORTER, PROBABLE ATP-DEPENDENT RNA HELICASE DDX6 | | Authors: | Basquin, J, Oezguer, S, Conti, E. | | Deposit date: | 2015-09-08 | | Release date: | 2015-10-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Structure of a Human 4E-T/Ddx6/Cnot1 Complex Reveals the Different Interplay of Ddx6-Binding Proteins with the Ccr4-not Complex.
Cell Rep., 13, 2015
|
|
7SAW
 
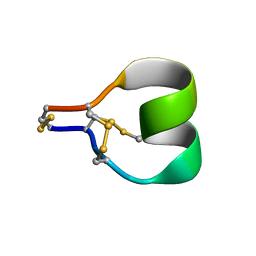 | | Mu-conotoxin KIIIA isomer 2 | | Descriptor: | Mu-conotoxin KIIIA | | Authors: | Schroeder, C.I, Tran, H.N.T. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into the inhibition of human voltage-gated sodium channels by mu-conotoxin KIIIA disulfide isomers.
J.Biol.Chem., 298, 2022
|
|
7VOI
 
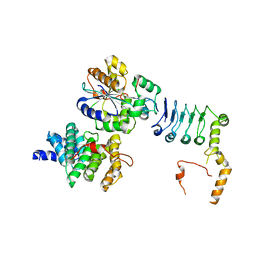 | | Structure of the human CNOT1(MIF4G)-CNOT6L-CNOT7 complex | | Descriptor: | CCR4-NOT transcription complex subunit 1, CCR4-NOT transcription complex subunit 6-like, CCR4-NOT transcription complex subunit 7 | | Authors: | Bartlam, M, Zhang, Q. | | Deposit date: | 2021-10-13 | | Release date: | 2022-06-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (4.38 Å) | | Cite: | Structure of the human Ccr4-Not nuclease module using X-ray crystallography and electron paramagnetic resonance spectroscopy distance measurements.
Protein Sci., 31, 2022
|
|
4GML
 
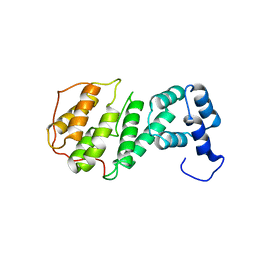 | |
4CQO
 
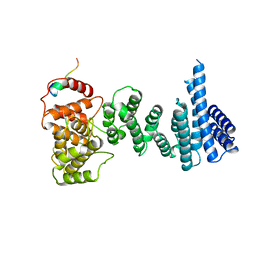 | | Structure of the human CNOT1 superfamily homology domain in complex with a Nanos1 peptide | | Descriptor: | CCR4-NOT TRANSCRIPTION COMPLEX SUBUNIT 1, NANOS HOMOLOG 1 | | Authors: | Raisch, T, Jonas, S, Weichenrieder, O, Bhandari, D, Izaurralde, E. | | Deposit date: | 2014-02-21 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis for the Nanos-Mediated Recruitment of the Ccr4-not Complex and Translational Repression
Genes Dev., 28, 2014
|
|
4J8S
 
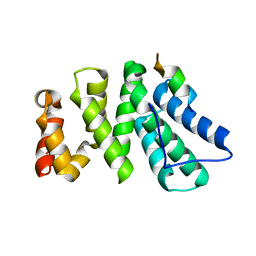 | | Crystal structure of human CNOT1 MIF4G domain in complex with a TTP peptide | | Descriptor: | CCR4-NOT transcription complex subunit 1, Tristetraprolin | | Authors: | Frank, F, Fabian, M.R, Rouya, C, Siddiqui, N, Lai, W.S, Karetnikov, A, Blackshear, P.J, Sonenberg, N, Nagar, B. | | Deposit date: | 2013-02-14 | | Release date: | 2013-05-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural basis for the recruitment of the human CCR4-NOT deadenylase complex by tristetraprolin.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3P1G
 
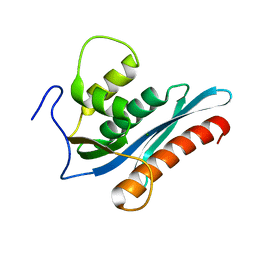 | |
6PY8
 
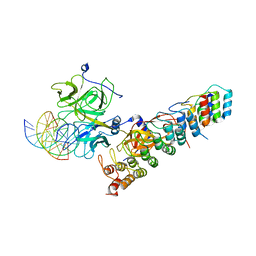 | | Crystal structure of the RBPJ-NOTCH1-NRARP ternary complex bound to DNA | | Descriptor: | DNA, Neurogenic locus notch homolog protein 1, Notch-regulated ankyrin repeat-containing protein, ... | | Authors: | Jarrett, S.M, Seegar, T.C.M, Blacklow, S.C. | | Deposit date: | 2019-07-29 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.75 Å) | | Cite: | Extension of the Notch intracellular domain ankyrin repeat stack by NRARP promotes feedback inhibition of Notch signaling.
Sci.Signal., 12, 2019
|
|
6WQU
 
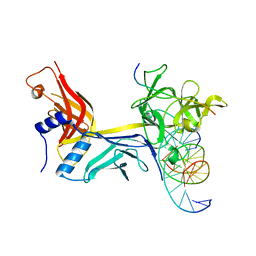 | | CSL (RBPJ) bound to Notch3 RAM and DNA | | Descriptor: | DNA (5'-D(*AP*AP*TP*CP*TP*TP*TP*CP*CP*CP*AP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*TP*AP*CP*CP*GP*TP*GP*GP*GP*AP*AP*AP*GP*A)-3'), Neurogenic locus notch homolog protein 3, ... | | Authors: | Kovall, R.A, Gagliani, E, Hall, D. | | Deposit date: | 2020-04-29 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | PIM-induced phosphorylation of Notch3 promotes breast cancer tumorigenicity in a CSL-independent fashion.
J.Biol.Chem., 296, 2021
|
|
5KY4
 
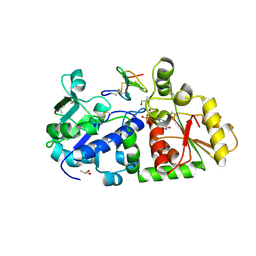 | | mouse POFUT1 in complex with mouse Notch1 EGF26 and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-21 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.472 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
5KY9
 
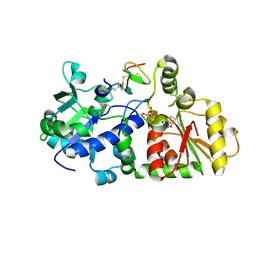 | |
5KY0
 
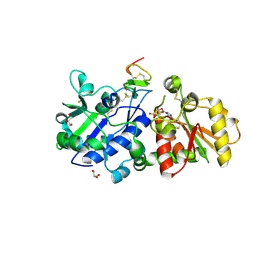 | | mouse POFUT1 in complex with mouse Notch1 EGF12(D464G) and GDP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GDP-fucose protein O-fucosyltransferase 1, GLYCEROL, ... | | Authors: | Li, Z, Rini, J.M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-05-17 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Recognition of EGF-like domains by the Notch-modifying O-fucosyltransferase POFUT1.
Nat. Chem. Biol., 13, 2017
|
|
5KY8
 
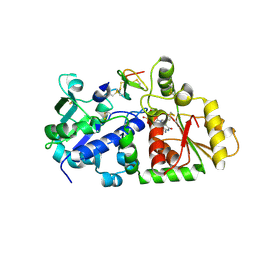 | |
5MWB
 
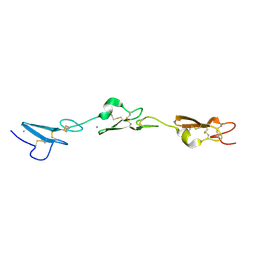 | | Human Notch-2 EGF11-13 | | Descriptor: | CALCIUM ION, Neurogenic locus notch homolog protein 2, alpha-L-fucopyranose, ... | | Authors: | Suckling, R.J, Handford, P.A, Lea, S.M. | | Deposit date: | 2017-01-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and functional dissection of the interplay between lipid and Notch binding by human Notch ligands.
EMBO J., 36, 2017
|
|
7ALJ
 
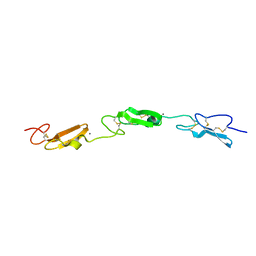 | | Structure of Drosophila Notch EGF domains 11-13 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neurogenic locus Notch protein, ... | | Authors: | Suckling, R, Johnson, S, Lea, S.M. | | Deposit date: | 2020-10-06 | | Release date: | 2021-08-04 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | The conserved C2 phospholipid-binding domain in Delta contributes to robust Notch signalling.
Embo Rep., 22, 2021
|
|
1EYO
 
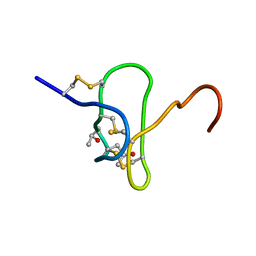 | |
1OT8
 
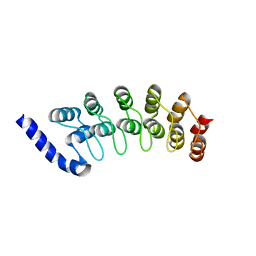 | | Structure of the Ankyrin Domain of the Drosophila Notch Receptor | | Descriptor: | MAGNESIUM ION, Neurogenic locus Notch protein | | Authors: | Zweifel, M.E, Leahy, D.J, Hughson, F.M, Barrick, D. | | Deposit date: | 2003-03-21 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and stability of the ankyrin domain of the Drosophila Notch receptor
Protein Sci., 12, 2003
|
|
1TTK
 
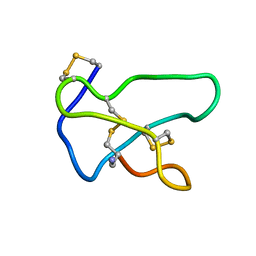 | | NMR solution structure of omega-conotoxin MVIIA, a N-type calcium channel blocker | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-22 | | Release date: | 2004-07-06 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
1TT3
 
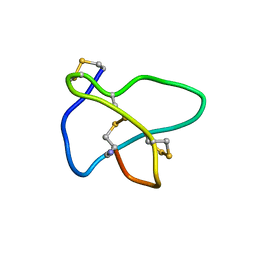 | | NMR soulution structure of omega-conotoxin [K10]MVIIA | | Descriptor: | Omega-conotoxin MVIIa | | Authors: | Adams, D.J, Smith, A.B, Schroeder, C.I, Yasuda, T, Lewis, R.J. | | Deposit date: | 2004-06-21 | | Release date: | 2004-07-06 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | omega-conotoxin CVID inhibits a pharmacologically distinct voltage-sensitive calcium channel associated with transmitter release from preganglionic nerve terminals
J.Biol.Chem., 278, 2003
|
|
6YUW
 
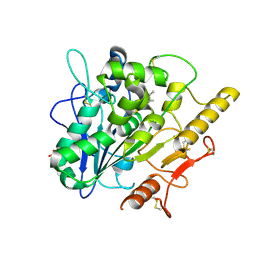 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLE-3-CARBOXYLIC ACID FRAGMENT 454 | | Descriptor: | 1-(cyclopropylmethyl)-2,5-dimethyl-pyrrole-3-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, Palmitoleoyl-protein carboxylesterase NOTUM, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
6YV2
 
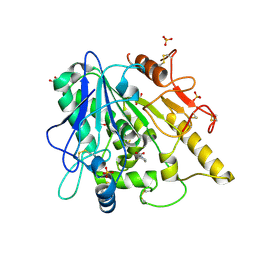 | | STRUCTURE OF THE WNT DEACYLASE NOTUM IN COMPLEX WITH A PYRROLIDINE-3-CARBOXYLIC ACID FRAGMENT 598 | | Descriptor: | (3~{R})-1-phenylpyrrolidine-3-carboxylic acid, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ruza, R.R, Hillier, J, Jones, E.Y. | | Deposit date: | 2020-04-27 | | Release date: | 2020-05-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Screening of a Custom-Designed Acid Fragment Library Identifies 1-Phenylpyrroles and 1-Phenylpyrrolidines as Inhibitors of Notum Carboxylesterase Activity.
J.Med.Chem., 63, 2020
|
|
