1ZSH
 
 | | Crystal structure of bovine arrestin-2 in complex with inositol hexakisphosphate (IP6) | | Descriptor: | Beta-arrestin 1, INOSITOL HEXAKISPHOSPHATE, MAGNESIUM ION | | Authors: | Milano, S.K, Kim, Y.M, Stefano, F.P, Benovic, J.L, Brenner, C. | | Deposit date: | 2005-05-24 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding
J.Biol.Chem., 281, 2006
|
|
1KDQ
 
 | | Crystal Structure Analysis of the Mutant S189D Rat Chymotrypsin | | Descriptor: | CALCIUM ION, CHYMOTRYPSIN B, B CHAIN, ... | | Authors: | Szabo, E, Bocskei, Z, Naray-Szabo, G, Graf, L, Venekei, I. | | Deposit date: | 2001-11-13 | | Release date: | 2003-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Three Dimensional Structures of S189D Chymotrypsin and D189S Trypsin Mutants: The Effect of Polarity at Site 189 on a Protease-specific Stabilization of
the Substrate-binding Site
J.Mol.Biol., 331, 2003
|
|
1JSY
 
 | | Crystal structure of bovine arrestin-2 | | Descriptor: | Bovine arrestin-2 (full length) | | Authors: | Milano, S.K, Pace, H.C, Kim, Y.M, Brenner, C, Benovic, J.L. | | Deposit date: | 2001-08-19 | | Release date: | 2002-03-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis.
Biochemistry, 41, 2002
|
|
6LA0
 
 | | Crystal structure of AoRut | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoside hydrolase family 5 | | Authors: | Koseki, T, Makabe, K. | | Deposit date: | 2019-11-11 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Aspergillus oryzae Rutinosidase: Biochemical and Structural Investigation.
Appl.Environ.Microbiol., 87, 2021
|
|
8AS2
 
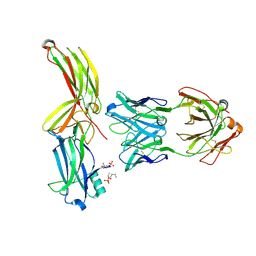 | | Structure of arrestin2 in complex with 4P CCR5 phosphopeptide and Fab30 | | Descriptor: | Beta-arrestin-1, C-C chemokine receptor type 5, Fab30 heavy chain, ... | | Authors: | Isaikina, P, Jakob, R.P, Maier, T, Grzesiek, S. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A key GPCR phosphorylation motif discovered in arrestin2⋅CCR5 phosphopeptide complexes.
Mol.Cell, 83, 2023
|
|
8AS3
 
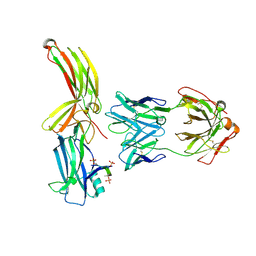 | | Structure of arrestin2 in complex with 6P CCR5 phosphopeptide and Fab30 | | Descriptor: | Beta-arrestin-1, C-C chemokine receptor type 5, Fab30 heavy chain, ... | | Authors: | Isaikina, P, Jakob, R.P, Maier, T, Grzesiek, S. | | Deposit date: | 2022-08-18 | | Release date: | 2023-06-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | A key GPCR phosphorylation motif discovered in arrestin2⋅CCR5 phosphopeptide complexes.
Mol.Cell, 83, 2023
|
|
8AS4
 
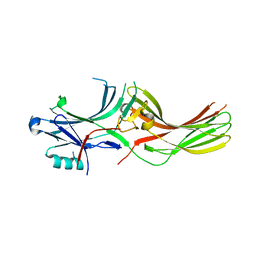 | |
1DEP
 
 | | MEMBRANE PROTEIN, NMR, 1 STRUCTURE | | Descriptor: | T345-359 | | Authors: | Jung, H, Schnackerz, K.D. | | Deposit date: | 1995-08-23 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR and circular dichroism studies of synthetic peptides derived from the third intracellular loop of the beta-adrenoceptor.
FEBS Lett., 358, 1995
|
|
6KL7
 
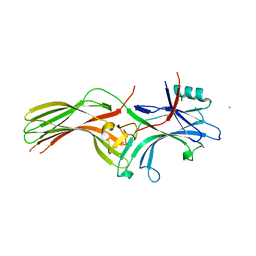 | | Beta-arrestin 1 mutant S13D/T275D | | Descriptor: | 1,2-ETHANEDIOL, BARIUM ION, Beta-arrestin-1 | | Authors: | Kang, H, Choi, H.J. | | Deposit date: | 2019-07-29 | | Release date: | 2020-01-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | Conformational Dynamics and Functional Implications of Phosphorylated beta-Arrestins.
Structure, 28, 2020
|
|
8I0Q
 
 | | Structure of beta-arrestin1 in complex with a phosphopeptide corresponding to the human C-X-C chemokine receptor type 4, CXCR4 (Local refine) | | Descriptor: | Beta-arrestin-1, C-X-C chemokine receptor type 4, Fab30 Heavy Chain, ... | | Authors: | Maharana, J, Sarma, P, Yadav, M.K, Banerjee, R, Shukla, A.K. | | Deposit date: | 2023-01-11 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.45 Å) | | Cite: | Structural snapshots uncover a key phosphorylation motif in GPCRs driving beta-arrestin activation.
Mol.Cell, 83, 2023
|
|
8HST
 
 | |
5MGR
 
 | | Human receptor NKR-P1 in glycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
2C07
 
 | | Oxoacyl-ACP reductase of Plasmodium falciparum | | Descriptor: | 3-OXOACYL-(ACYL-CARRIER PROTEIN) REDUCTASE, SULFATE ION | | Authors: | Urch, J.E, Wickramasinghe, S.R, Inglis, K.A, Muller, S, Fairlamb, A.H, van Aalten, D.M.F. | | Deposit date: | 2005-08-26 | | Release date: | 2005-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Kinetic, Inhibition and Structural Studies on 3-Oxoacyl-Acp Reductase from Plasmodium Falciparum, a Key Enzyme in Fatty Acid Biosynthesis.
Biochem.J., 393, 2006
|
|
5MGS
 
 | | Human receptor NKR-P1 in deglycosylated form, extracellular domain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Killer cell lectin-like receptor subfamily B member 1 | | Authors: | Skalova, T, Blaha, J, Stransky, J, Koval, T, Hasek, J, Yuguang, Z, Harlos, K, Vanek, O, Dohnalek, J. | | Deposit date: | 2016-11-22 | | Release date: | 2018-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the human NK cell NKR-P1:LLT1 receptor:ligand complex reveals clustering in the immune synapse.
Nat Commun, 13, 2022
|
|
6NI2
 
 | | Stabilized beta-arrestin 1-V2T subcomplex of a GPCR-G protein-beta-arrestin mega-complex | | Descriptor: | Beta-arrestin-1, Fab30 Heavy Chain, Fab30 Light Chain, ... | | Authors: | Nguyen, A.H, Thomsen, A.R.B, Cahill, T.J, des Georges, A, Lefkowitz, R.J. | | Deposit date: | 2018-12-26 | | Release date: | 2019-11-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure of an endosomal signaling GPCR-G protein-beta-arrestin megacomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6UP7
 
 | | neurotensin receptor and arrestin2 complex | | Descriptor: | ARG-ARG-PRO-TYR-ILE-LEU, Beta-arrestin-1, Neurotensin receptor type 1, ... | | Authors: | Qu, Q.H, Huang, W, Masureel, M, Janetzko, J, Kobilka, B.K, Skiniotis, G. | | Deposit date: | 2019-10-16 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of the neurotensin receptor 1 in complex with beta-arrestin 1.
Nature, 579, 2020
|
|
4RCM
 
 | | Crystal structure of the Pho92 YTH domain in complex with m6A | | Descriptor: | Methylated RNA-binding protein 1, RNA (5'-R(*UP*G)-D(*(6MZ)P*CP*U)-3'), UNKNOWN ATOM OR ION | | Authors: | Tempel, W, Xu, C, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-09-16 | | Release date: | 2014-11-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for the Discriminative Recognition of N6-Methyladenosine RNA by the Human YT521-B Homology Domain Family of Proteins.
J.Biol.Chem., 290, 2015
|
|
6H7N
 
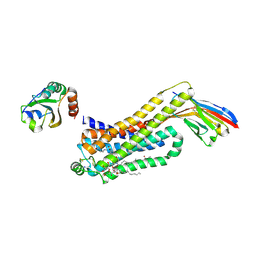 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND PARTIAL AGONIST XAMOTEROL AND NANOBODY Nb6B9 | | Descriptor: | Beta-1 adrenergic receptor, Camelid antibody fragment Nb6B9, HEGA-10, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis for high affinity agonist binding in GPCRs
Biorxiv, 2018
|
|
6H7M
 
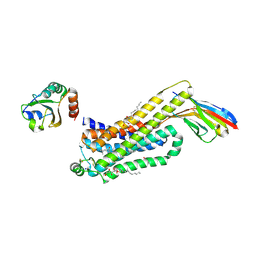 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND PARTIAL AGONIST SALBUTAMOL AND NANOBODY Nb6B9 | | Descriptor: | Beta-1 adrenergic receptor, Camelid antibody fragment Nb6B9, HEGA-10, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Molecular basis for high-affinity agonist binding in GPCRs.
Science, 364, 2019
|
|
8PPM
 
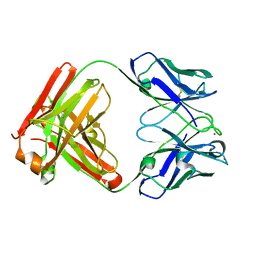 | | IL-12Rb1 neutralizing Fab4, crystal kappa variant | | Descriptor: | Fab4 Light chain, crystal kappa variant, Fab4 heavy chain, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-07-07 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of complete extracellular receptor assemblies mediated by IL-12 and IL-23.
Nat.Struct.Mol.Biol., 31, 2024
|
|
6IBL
 
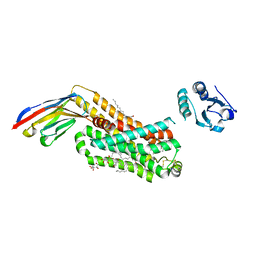 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND AGONIST FORMOTEROL AND NANOBODY Nb80 | | Descriptor: | Camelid antibody fragment Nb80, HEGA-10, SODIUM ION, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis of beta-arrestin coupling to formoterol-bound beta1-adrenoceptor.
Nature, 583, 2020
|
|
6H7L
 
 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND PARTIAL AGONIST DOBUTAMINE AND NANOBODY Nb6B9 | | Descriptor: | Beta-1 adrenergic receptor, Camelid antibody fragment Nb6B9, DOBUTAMINE, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Molecular basis for high-affinity agonist binding in GPCRs.
Science, 364, 2019
|
|
6H7O
 
 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND WEAK PARTIAL AGONIST CYANOPINDOLOL AND NANOBODY Nb6B9 | | Descriptor: | 4-{[(2S)-3-(tert-butylamino)-2-hydroxypropyl]oxy}-3H-indole-2-carbonitrile, Beta-1 adrenergic receptor, Camelid antibody fragment Nb6B9, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for high-affinity agonist binding in GPCRs.
Science, 364, 2019
|
|
6H7J
 
 | | ACTIVATED TURKEY BETA1 ADRENOCEPTOR WITH BOUND AGONIST ISOPRENALINE AND NANOBODY Nb80 | | Descriptor: | Beta-1 adrenergic receptor, Camelid antibody fragment Nb80, HEGA-10, ... | | Authors: | Warne, T, Edwards, P.C, Dore, A.S, Leslie, A.G.W, Tate, C.G. | | Deposit date: | 2018-07-31 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for high-affinity agonist binding in GPCRs.
Science, 364, 2019
|
|
1G4M
 
 | | CRYSTAL STRUCTURE OF BOVINE BETA-ARRESTIN 1 | | Descriptor: | BETA-ARRESTIN1 | | Authors: | Schubert, C, Han, M. | | Deposit date: | 2000-10-27 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane Translocation.
Structure, 9, 2001
|
|
