5FV8
 
 | |
6YA4
 
 | |
7KYM
 
 | |
6HBV
 
 | |
6FFY
 
 | | Structure of the mouse SorCS2-NGF complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-nerve growth factor, VPS10 domain-containing receptor SorCS2, ... | | Authors: | Leloup, N.O.L, Janssen, B.J.C. | | Deposit date: | 2018-01-09 | | Release date: | 2018-08-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural insights into SorCS2-Nerve Growth Factor complex formation.
Nat Commun, 9, 2018
|
|
6CNP
 
 | |
6YA3
 
 | |
6YAB
 
 | |
6HJ3
 
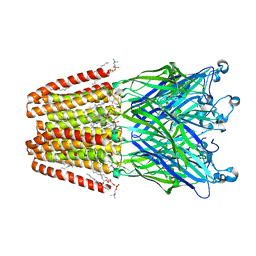 | | Xray structure of GLIC in complex with fumarate | | Descriptor: | CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Fourati, Z, Delarue, M. | | Deposit date: | 2018-08-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural evidence for the binding of monocarboxylates and dicarboxylates at pharmacologically relevant extracellular sites of a pentameric ligand-gated ion channel.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6CK2
 
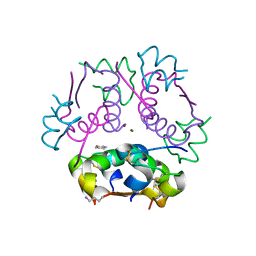 | | Insulin analog containing a YB26W mutation | | Descriptor: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | Authors: | Rege, N.K, Yee, V.C, Weiss, M.A. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based stabilization of insulin as a therapeutic protein assembly via enhanced aromatic-aromatic interactions.
J. Biol. Chem., 293, 2018
|
|
8A9W
 
 | | Crystal structure of PulL C-ter domain | | Descriptor: | SULFATE ION, Type II secretion system protein L | | Authors: | Dazzoni, R, Li, Y, Lopez-Castilla, A, Brier, S, Mechaly, A, Cordier, F, Haouz, A, Nilges, M, Francetic, O, Bardiaux, B, Izadi-Pruneyre, N. | | Deposit date: | 2022-06-29 | | Release date: | 2023-01-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | Structure and dynamic association of an assembly platform subcomplex of the bacterial type II secretion system.
Structure, 31, 2023
|
|
5TWZ
 
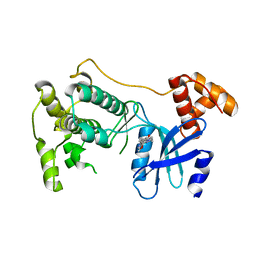 | | Structure of Maternal Embryonic Leucine Zipper Kinase | | Descriptor: | 7-{[2-methoxy-4-(1H-pyrazol-4-yl)benzoyl]amino}-2,3,4,5-tetrahydro-1H-3-benzazepinium, Maternal embryonic leucine zipper kinase | | Authors: | Seo, H.-Y, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | MELK is not necessary for the proliferation of basal-like breast cancer cells.
Elife, 6, 2017
|
|
5FO8
 
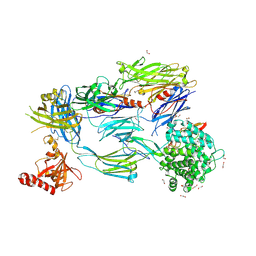 | | Crystal Structure of Human Complement C3b in Complex with MCP (CCP1-4) | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
7QK9
 
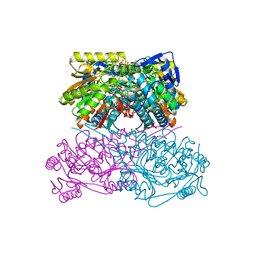 | | Crystal structure of the ALDH1A3-ATP complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aldehyde dehydrogenase family 1 member A3, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Castellvi, A, Farres, J. | | Deposit date: | 2021-12-17 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.782 Å) | | Cite: | Structural and biochemical evidence that ATP inhibits the cancer biomarker human aldehyde dehydrogenase 1A3.
Commun Biol, 5, 2022
|
|
6PZO
 
 | |
6HJK
 
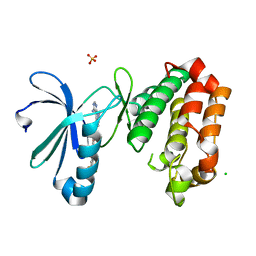 | | Crystal Structure of Aurora-A L210C catalytic domain in complex with ASDO2 | | Descriptor: | (~{E})-~{N}-[4-(4-azanyl-1-propan-2-yl-pyrazolo[3,4-d]pyrimidin-3-yl)phenyl]-4-[4-fluoranyl-3-(trifluoromethyl)phenyl]-4-oxidanylidene-but-2-enamide, Aurora kinase A, CHLORIDE ION, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2018-09-04 | | Release date: | 2018-10-03 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Type II Kinase Inhibitors Targeting Cys-Gatekeeper Kinases Display Orthogonality with Wild Type and Ala/Gly-Gatekeeper Kinases.
ACS Chem. Biol., 13, 2018
|
|
6YQ9
 
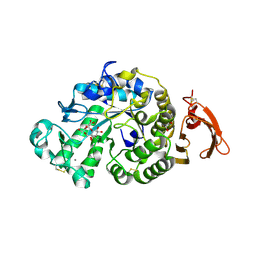 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol epoxide inhibitor | | Descriptor: | (1R,2R,3S,5R,6S)-2,3,5-trihydroxy-6-(hydroxymethyl)cyclohexyl alpha-D-glucopyranoside, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
5FWU
 
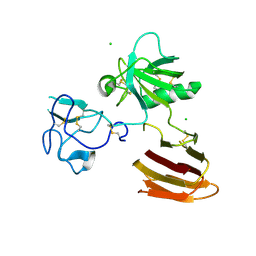 | | Wnt modulator Kremen crystal form II at 2.8A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, KREMEN PROTEIN 1 | | Authors: | Zebisch, M, Jackson, V.A, Jones, E.Y. | | Deposit date: | 2016-02-21 | | Release date: | 2016-07-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Dual-Mode Wnt Regulator Kremen1 and Insight Into Ternary Complex Formation with Lrp6 and Dickkopf
Structure, 24, 2016
|
|
6PZU
 
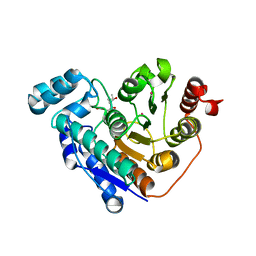 | |
6YQC
 
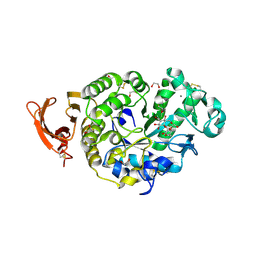 | | Taka-amylase in complex with alpha-glucosyl epi-cyclophellitol epoxide inhibitor | | Descriptor: | (1~{R},2~{S},4~{R},5~{S},6~{R})-6-[(2~{S},3~{R},4~{R},5~{S},6~{R})-5-heptoxy-6-(hydroxymethyl)-3,4-bis(oxidanyl)oxan-2-yl]oxy-5-(hydroxymethyl)cyclohexane-1,2,4-triol, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Armstrong, Z, Chen, Y, Artola, M, Overkleeft, H, Davies, G. | | Deposit date: | 2020-04-16 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Activity-Based Protein Profiling of Retaining alpha-Amylases in Complex Biological Samples.
J.Am.Chem.Soc., 143, 2021
|
|
6HJD
 
 | | Cholera toxin classical B-pentamer in complex with Lewis-x | | Descriptor: | BICINE, CALCIUM ION, Cholera toxin B subunit, ... | | Authors: | Krengel, U, Heim, J.B. | | Deposit date: | 2018-09-03 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Crystal structures of cholera toxin in complex with fucosylated receptors point to importance of secondary binding site.
Sci Rep, 9, 2019
|
|
6FJ8
 
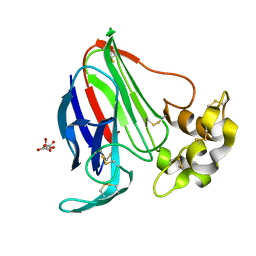 | |
6FHA
 
 | |
6Y7V
 
 | | Crystal structure of the KDEL receptor bound to HDEL peptide at pH 6.0 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON DIOXIDE, ER lumen protein-retaining receptor 2, ... | | Authors: | Braeuer, P, Newstead, S. | | Deposit date: | 2020-03-02 | | Release date: | 2021-02-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.241 Å) | | Cite: | A signal capture and proofreading mechanism for the KDEL-receptor explains selectivity and dynamic range in ER retrieval.
Elife, 10, 2021
|
|
6HJS
 
 | | Crystal structure of glutathione transferase Omega 1C from Trametes versicolor | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLUTATHIONE, Uncharacterized protein | | Authors: | Schwartz, M, Favier, F, Didierjean, C. | | Deposit date: | 2018-09-04 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.612 Å) | | Cite: | Diversity of Omega Glutathione Transferases in mushroom-forming fungi revealed by phylogenetic, transcriptomic, biochemical and structural approaches.
Fungal Genet Biol., 2021
|
|
