1IYY
 
 | | NMR STRUCTURE OF Gln25-RIBONUCLEASE T1, 24 STRUCTURES | | Descriptor: | RIBONUCLEASE T1 | | Authors: | Hatano, K, Kojima, M, Suzuki, E, Tanokura, M, Takahashi, K. | | Deposit date: | 2002-09-12 | | Release date: | 2003-10-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Determination of the NMR structure of Gln25-ribonuclease T1.
Biol. Chem., 384, 2003
|
|
6I7B
 
 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 3. | | Descriptor: | Nucleoprotein | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Carlero, D, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-11-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
5WG7
 
 | | Human Carbonic Anhydrase II complexed with AceK | | Descriptor: | Acesulfame, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Murray, A.B, Lomelino, C.L, Supuran, C.T, McKenna, R. | | Deposit date: | 2017-07-13 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | "Seriously Sweet": Acesulfame K Exhibits Selective Inhibition Using Alternative Binding Modes in Carbonic Anhydrase Isoforms.
J. Med. Chem., 61, 2018
|
|
5WO4
 
 | | JAK1 complexed with compound 28 | | Descriptor: | 3-[(4-chloro-3-methoxyphenyl)amino]-1-[(3R,4S)-4-cyanooxan-3-yl]-1H-pyrazole-4-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lesburg, C.A, Patel, S.B. | | Deposit date: | 2017-08-01 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Discovery of 3-((4-Chloro-3-methoxyphenyl)amino)-1-((3R,4S)-4-cyanotetrahydro-2H-pyran-3-yl)-1H-pyrazole-4-carboxamide, a Highly Ligand Efficient and Efficacious Janus Kinase 1 Selective Inhibitor with Favorable Pharmacokinetic Properties.
J. Med. Chem., 60, 2017
|
|
7TBM
 
 | | Composite structure of the dilated human nuclear pore complex (NPC) generated with a 37A in situ cryo-ET map of CD4+ T cell NPC | | Descriptor: | DDX19, NUP107 CTD, NUP107 NTD, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-15 | | Last modified: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (37 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
3IGV
 
 | | Crystal structure of HCV NS5B polymerase with a novel monocyclic dihydro-pyridinone inhibitor | | Descriptor: | N-{3-[(6S)-6-ethyl-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-1,2,5,6-tetrahydropyridin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2009-07-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 5,5'- and 6,6'-dialkyl-5,6-dihydro-1H-pyridin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
8AG4
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Protein C10, X-ray repair cross-complementing protein 5, X-ray repair cross-complementing protein 6 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
8AG5
 
 | | Vaccinia C16 protein bound to Ku70/Ku80 | | Descriptor: | Ku70-Xrcc6, Protein C10, X-ray repair cross-complementing protein 5 | | Authors: | Rivera-Calzada, A, Arribas-Bosacoma, R, Pearl, L.H, Llorca, O. | | Deposit date: | 2022-07-19 | | Release date: | 2022-11-09 | | Last modified: | 2022-11-30 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Structural basis for the inactivation of cytosolic DNA sensing by the vaccinia virus.
Nat Commun, 13, 2022
|
|
8D96
 
 | | Human DNA polymerase alpha/primase elongation complex I bound to primer/template | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, DNA (5'-D(*AP*TP*AP*AP*TP*GP*GP*TP*CP*GP*TP*GP*CP*CP*GP*CP*CP*AP*AP*TP*AP*A)-3'), DNA polymerase alpha catalytic subunit, ... | | Authors: | He, Q, Baranovskiy, A, Lim, C, Tahirov, T. | | Deposit date: | 2022-06-09 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structures of human primosome elongation complexes.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5TRI
 
 | | CRYSTAL STRUCTURE OF THE HEPATITIS C VIRUS NS5B RNA-DEPENDENT RNA POLYMERASE IN COMPLEX WITH 3-[(4-chlorophenyl)methoxy]-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)benzoic acid | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, 3-[(4-chlorophenyl)methoxy]-2-(1-oxo-1,3-dihydro-2H-isoindol-2-yl)benzoic acid, NS5B RNA-DEPENDENT RNA POLYMERASE, ... | | Authors: | Sheriff, S. | | Deposit date: | 2016-10-26 | | Release date: | 2016-11-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and initial optimization of alkoxyanthranilic acid derivatives as inhibitors of HCV NS5B polymerase.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
2W2G
 
 | | Human SARS coronavirus unique domain | | Descriptor: | NON-STRUCTURAL PROTEIN 3, SULFATE ION | | Authors: | Tan, J, Vonrhein, C, Smart, O.S, Bricogne, G, Bollati, M, Kusov, Y, Hansen, G, Mesters, J.R, Schmidt, C.L, Hilgenfeld, R. | | Deposit date: | 2008-10-30 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The Sars-Unique Domain (Sud) of Sars Coronavirus Contains Two Macrodomains that Bind G-Quadruplexes.
Plos Pathog., 5, 2009
|
|
8ACB
 
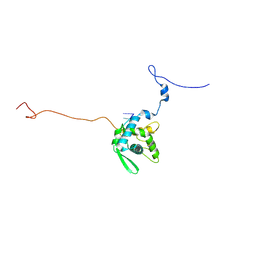 | | CryoEM structure of sweet potato feathery mottle virus VLP | | Descriptor: | Genome polyprotein, Single-stranded RNA | | Authors: | Javed, A, Byrne, J.M, Ranson, N, Lomonosoff, G. | | Deposit date: | 2022-07-05 | | Release date: | 2023-05-17 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | CryoEM and stability analysis of virus-like particles of potyvirus and ipomovirus infecting a common host.
Commun Biol, 6, 2023
|
|
8ACC
 
 | | CryoEM structure of sweet potato mild mottle virus VLP | | Descriptor: | Polyprotein, Single-stranded RNA | | Authors: | Javed, A, Byrne, B.M, Ranson, N, Lomonosoff, G. | | Deposit date: | 2022-07-05 | | Release date: | 2023-05-17 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | CryoEM and stability analysis of virus-like particles of potyvirus and ipomovirus infecting a common host.
Commun Biol, 6, 2023
|
|
6H9G
 
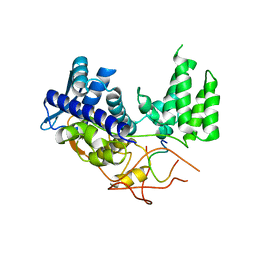 | | Influenza A nucleoprotein docked into 3D helical structure of the wild type ribonucleoprotein complex obtained using cryoEM. Conformation 1. | | Descriptor: | Nucleoprotein, Polypeptide loop | | Authors: | Coloma, R, Arranz, R, de la Rosa-Trevin, J.M, Sorzano, C.O.S, Munier, S, Carlero, D, Naffakh, N, Ortin, J, Martin-Benito, J. | | Deposit date: | 2018-08-03 | | Release date: | 2020-02-12 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11 Å) | | Cite: | Structural insights into influenza A virus ribonucleoproteins reveal a processive helical track as transcription mechanism.
Nat Microbiol, 5, 2020
|
|
8BMW
 
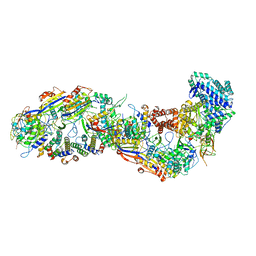 | | SsoCsm | | Descriptor: | CRISPR-associated Cas7 paralog (Type III-D), CRISPR-associated protein Cas10 (Type III-D), CRISPR-associated protein Cas5 (Type III-D), ... | | Authors: | Spagnolo, L, White, M.F. | | Deposit date: | 2022-11-11 | | Release date: | 2023-03-01 | | Last modified: | 2023-03-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Saccharolobus solfataricus type III-D CRISPR effector.
Curr Res Struct Biol, 5, 2023
|
|
7V2W
 
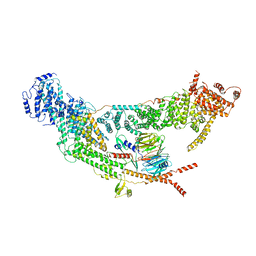 | | protomer structure from the dimer of yeast THO complex | | Descriptor: | Protein TEX1, THO complex subunit 2, THO complex subunit HPR1, ... | | Authors: | Chen, C, Tan, M, Wu, Z, Wu, J, Lei, M. | | Deposit date: | 2021-08-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional insights into R-loop prevention and mRNA export by budding yeast THO-Sub2 complex.
Sci Bull (Beijing), 66, 2021
|
|
5V8W
 
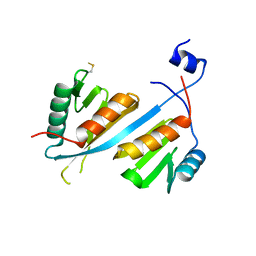 | | Crystal structure of human Integrator IntS9-IntS11 CTD complex | | Descriptor: | Integrator complex subunit 11, Integrator complex subunit 9 | | Authors: | Wu, Y, Tong, L. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-12 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular basis for the interaction between Integrator subunits IntS9 and IntS11 and its functional importance.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1GMP
 
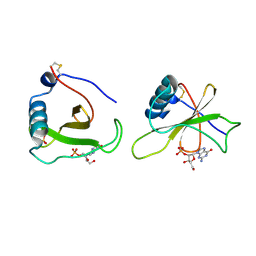 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1GMR
 
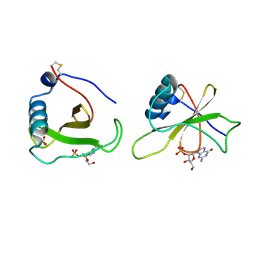 | | COMPLEX OF RIBONUCLEASE FROM STREPTOMYCES AUREOFACIENS WITH 2'-GMP AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | GUANOSINE-2'-MONOPHOSPHATE, RIBONUCLEASE SA, SULFATE ION | | Authors: | Sevcik, J, Hill, C, Dauter, Z, Wilson, K. | | Deposit date: | 1992-10-01 | | Release date: | 1993-10-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Complex of ribonuclease from Streptomyces aureofaciens with 2'-GMP at 1.7 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
4A4F
 
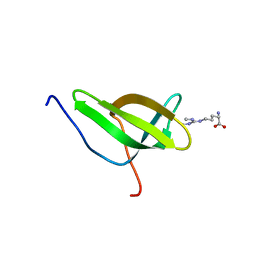 | | Solution structure of SPF30 Tudor domain in complex with symmetrically dimethylated arginine | | Descriptor: | N3, N4-DIMETHYLARGININE, SURVIVAL OF MOTOR NEURON-RELATED-SPLICING FACTOR 30 | | Authors: | Tripsianes, K, Madl, T, Machyna, M, Fessas, D, Englbrecht, C, Fischer, U, Neugebauer, K.M, Sattler, M. | | Deposit date: | 2011-10-12 | | Release date: | 2011-11-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for Dimethyl-Arginine Recognition by the Tudor Domains of Human Smn and Spf30 Proteins
Nat.Struct.Mol.Biol., 18, 2011
|
|
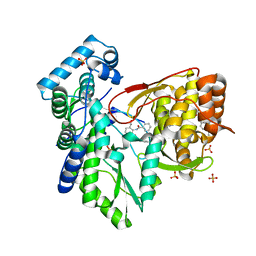
5TRH
 
 | |
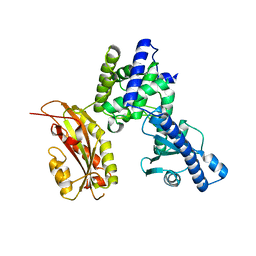
2O1P
 
 | |
6GYV
 
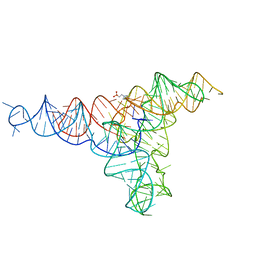 | | Lariat-capping ribozyme (circular permutation form) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lariat-capping ribozyme, MAGNESIUM ION, ... | | Authors: | Masquida, B, Meyer, M, Nielsen, H, Olieric, V, Roblin, P, Johansen, S.D, Westhof, E. | | Deposit date: | 2018-07-02 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.50003624 Å) | | Cite: | Speciation of a group I intron into a lariat capping ribozyme.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
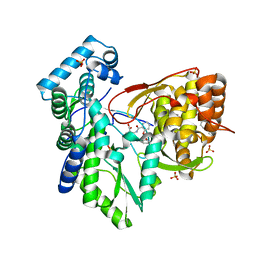
5TRJ
 
 | |
7AOI
 
 | | Trypanosoma brucei mitochondrial ribosome large subunit assembly intermediate | | Descriptor: | 50S ribosomal protein L13, 50S ribosomal protein L14, 50S ribosomal protein L17, ... | | Authors: | Tobiasson, V, Gahura, O, Aibara, S, Baradaran, R, Zikova, A, Amunts, A. | | Deposit date: | 2020-10-14 | | Release date: | 2020-12-02 | | Last modified: | 2021-03-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Interconnected assembly factors regulate the biogenesis of mitoribosomal large subunit.
Embo J., 40, 2021
|
|
