4U8H
 
 | | Crystal Structure of Mammalian Period-Cryptochrome Complex | | Descriptor: | Cryptochrome-2, Period circadian protein homolog 2, ZINC ION | | Authors: | Nangle, S.N, Rosensweig, C, Koike, N, Tei, H, Takahashi, J.S, Green, C.B, Zheng, N. | | Deposit date: | 2014-08-03 | | Release date: | 2014-10-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.798 Å) | | Cite: | Molecular assembly of the period-cryptochrome circadian transcriptional repressor complex.
Elife, 3, 2014
|
|
7NNR
 
 | | Crystal structure of Mycobacterium tuberculosis ArgC in complex with xanthene-9-carboxylic acid | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 9~{H}-xanthene-9-carboxylic acid, N-acetyl-gamma-glutamyl-phosphate reductase | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLR
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 2-phenyl-1H-imidazole | | Descriptor: | 2-phenyl-1H-imidazole, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
4UA4
 
 | |
7P4O
 
 | | Crystal structure of Autotaxin and 9(R)-delta6a,10a-THC | | Descriptor: | (9~{R},10~{a}~{S})-6,6,9-trimethyl-3-pentyl-6~{a},7,8,9,10,10~{a}-hexahydrobenzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A, Hausmann, J. | | Deposit date: | 2021-07-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Linking medicinal cannabis to autotaxin-lysophosphatidic acid signaling.
Life Sci Alliance, 6, 2023
|
|
4UAH
 
 | |
7NN8
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1H-indole-3-carbonitrile. | | Descriptor: | 1,2-ETHANEDIOL, 1~{H}-indole-3-carbonitrile, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.267 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NN4
 
 | | Crystal structure of Mycobacterium tuberculosis ArgD with prosthetic group pyridoxal 5'-phosphate and 3-hydroxy-2-naphthoic acid. | | Descriptor: | 3-hydroxynaphthalene-2-carboxylic acid, Acetylornithine aminotransferase, NITRATE ION | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
4UBO
 
 | | KINETIC CRYSTALLOGRAPHY OF ALPHA_E7-CARBOXYLESTERSE FROM LUCILLA CUPRINA - ABSORBED X-RAY DOSE 3.70 MGy TEMP 150K | | Descriptor: | DIETHYL HYDROGEN PHOSPHATE, E3 | | Authors: | Jackson, C.J, Carr, P.D, Weik, M, Huber, T, Meirelles, T, Correy, G. | | Deposit date: | 2014-08-13 | | Release date: | 2015-08-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Mapping the Accessible Conformational Landscape of an Insect Carboxylesterase Using Conformational Ensemble Analysis and Kinetic Crystallography
Structure, 24, 2016
|
|
7NLN
 
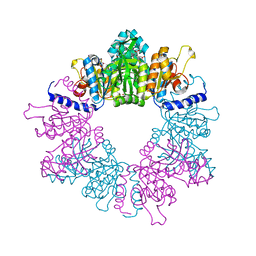 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with N-acetyl-glutamate | | Descriptor: | 1,2-ETHANEDIOL, Acetylglutamate kinase, N-ACETYL-L-GLUTAMATE | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLO
 
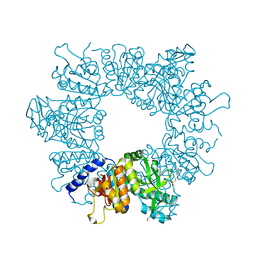 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with L-arginine | | Descriptor: | 1,2-ETHANEDIOL, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ARGININE, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
4UE3
 
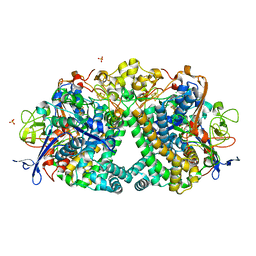 | | The Mechanism of Hydrogen Activation by NiFe-hydrogenases and the Importance of the active site Arginine | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, CHLORIDE ION, FE3-S4 CLUSTER, ... | | Authors: | Evans, R.M, Wehlin, S.A.M, Nomerotskaia, E, Sargent, F, Carr, S.B, Phillips, S.E.V, Armstrong, F.A. | | Deposit date: | 2014-12-15 | | Release date: | 2014-12-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mechanism of Hydrogen Activation by [Nife] Hydrogenases.
Nat.Chem.Biol., 12, 2016
|
|
7NN1
 
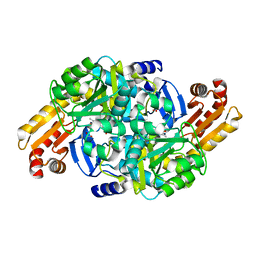 | | Crystal structure of Mycobacterium tuberculosis ArgD with prosthetic group pyridoxal 5'-phosphate | | Descriptor: | Acetylornithine aminotransferase, NITRATE ION | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NLZ
 
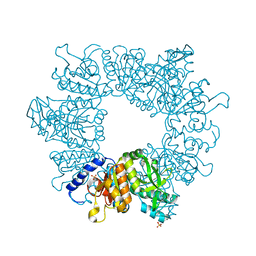 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 5-Methoxy-6-(trifluoromethyl)indole. | | Descriptor: | 5-methoxy-6-(trifluoromethyl)-1~{H}-indole, Acetylglutamate kinase, SULFATE ION | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-22 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.163 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7P4J
 
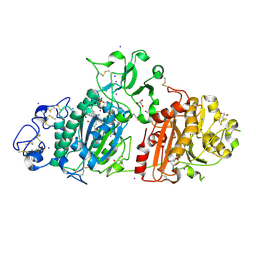 | | Crystal structure of Autotaxin and tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 7alpha-hydroxycholesterol, ... | | Authors: | Eymery, M.C, McCarthy, A.A, Hausmann, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Linking medicinal cannabis to autotaxin-lysophosphatidic acid signaling.
Life Sci Alliance, 6, 2023
|
|
7NPJ
 
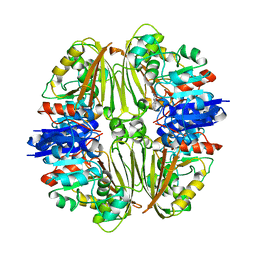 | | Crystal structure of Mycobacterium tuberculosis ArgC in complex with 6-phenoxy-3-pyridinamine | | Descriptor: | 6-phenoxy-3-pyridinamine, N-acetyl-gamma-glutamyl-phosphate reductase | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-27 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNC
 
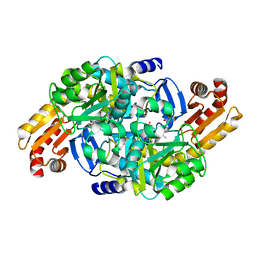 | | Crystal structure of Mycobacterium tuberculosis ArgD with prosthetic group pyridoxal-5'-phosphate and 6-Methoxyquinoline-3-carboxylic acid | | Descriptor: | 6-methoxyquinoline-3-carboxylic acid, Acetylornithine aminotransferase, NITRATE ION | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-24 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7N7I
 
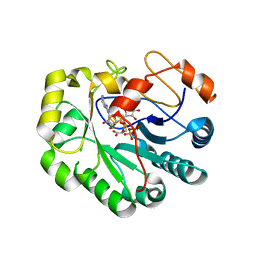 | | X-ray crystal structure of Viperin-like enzyme from Trichoderma virens | | Descriptor: | IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE, URIDINE 5'-TRIPHOSPHATE, ... | | Authors: | Grove, T.L, Almo, S.C, Bonanno, J.B, Lachowicz, J.C, Gizzi, A.G. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structural Insight into the Substrate Scope of Viperin and Viperin-like Enzymes from Three Domains of Life.
Biochemistry, 60, 2021
|
|
4U3J
 
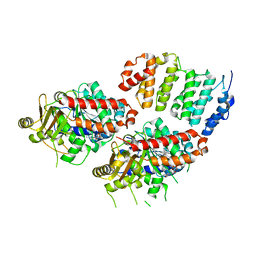 | | TOG2:alpha/beta-tubulin complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Protein STU2, ... | | Authors: | Ayaz, P, Rice, L.M. | | Deposit date: | 2014-07-22 | | Release date: | 2014-08-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A tethered delivery mechanism explains the catalytic action of a microtubule polymerase.
Elife, 3, 2014
|
|
1AKM
 
 | |
4U3T
 
 | |
7N7M
 
 | | Crystal Structure of PI5P4KIIAlpha complex with BI-2536 | | Descriptor: | 4-{[(7R)-8-cyclopentyl-7-ethyl-5-methyl-6-oxo-5,6,7,8-tetrahydropteridin-2-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Phosphatidylinositol 5-phosphate 4-kinase type-2 alpha, SULFATE ION | | Authors: | Chen, S, Ha, Y. | | Deposit date: | 2021-06-10 | | Release date: | 2021-06-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Pharmacological inhibition of PI5P4K alpha / beta disrupts cell energy metabolism and selectively kills p53-null tumor cells.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NM0
 
 | | Crystal structure of Mycobacterium tuberculosis ArgB in complex with 1-(2,6-dihydroxyphenyl)ethan-1-one. | | Descriptor: | 1,2-ETHANEDIOL, 1-[2,6-bis(oxidanyl)phenyl]ethanone, Acetylglutamate kinase, ... | | Authors: | Mendes, V, Thomas, S.E, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2021-02-23 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.281 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NOT
 
 | | Crystal structure of Mycobacterium tuberculosis ArgC in complex with nicotinamide adenine dinucleotide phosphate (NADP+) and 5-Methoxy-3-indoleacetic acid | | Descriptor: | (5-methoxy-1H-indol-3-yl)acetic acid, N-acetyl-gamma-glutamyl-phosphate reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
7NNI
 
 | |
