2D2C
 
 | | Crystal Structure Of Cytochrome B6F Complex with DBMIB From M. Laminosus | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 2,5-DIBROMO-3-ISOPROPYL-6-METHYLBENZO-1,4-QUINONE, Apocytochrome f, ... | | Authors: | Yan, J, Kurisu, G, Cramer, W.A. | | Deposit date: | 2005-09-07 | | Release date: | 2005-12-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Intraprotein transfer of the quinone analogue inhibitor 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone in the cytochrome b6f complex
Proc.Natl.Acad.Sci.USA, 103, 2006
|
|
3P80
 
 | |
2RAA
 
 | |
3E0B
 
 | | Bacillus anthracis Dihydrofolate Reductase complexed with NADPH and 2,4-diamino-5-(3-(2,5-dimethoxyphenyl)prop-1-ynyl)-6-ethylpyrimidine (UCP120B) | | Descriptor: | 5-[3-(2,5-dimethoxyphenyl)prop-1-yn-1-yl]-6-ethylpyrimidine-2,4-diamine, Dihydrofolate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Anderson, A.C, Beierlein, J.M, Frey, K.M. | | Deposit date: | 2008-07-31 | | Release date: | 2008-11-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Synthetic and Crystallographic Studies of a New Inhibitor Series Targeting Bacillus anthracis Dihydrofolate Reductase
J.Med.Chem., 51, 2008
|
|
4E96
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor PFi-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-N-(3-methyl-2-oxo-1,2,3,4-tetrahydroquinazolin-6-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Fish, P, Bunnage, M, Owen, D, Knapp, S, Cook, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of a chemical probe for bromo and extra C-terminal bromodomain inhibition through optimization of a fragment-derived hit.
J.Med.Chem., 55, 2012
|
|
3UDU
 
 | | Crystal structure of putative 3-isopropylmalate dehydrogenase from Campylobacter jejuni | | Descriptor: | 1,2-ETHANEDIOL, 3-isopropylmalate dehydrogenase, CHLORIDE ION | | Authors: | Tkaczuk, K.L, Chruszcz, M, Grimshaw, S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-10-28 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of putative 3-isopropylmalate dehydrogenase from Campylobacter jejuni
To be Published
|
|
1I0Z
 
 | | HUMAN HEART L-LACTATE DEHYDROGENASE H CHAIN, TERNARY COMPLEX WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-LACTATE DEHYDROGENASE H CHAIN, OXAMIC ACID | | Authors: | Read, J.A, Winter, V.J, Eszes, C.M, Sessions, R.B, Brady, R.L. | | Deposit date: | 2001-01-30 | | Release date: | 2001-03-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for altered activity of M- and H-isozyme forms of human lactate dehydrogenase.
Proteins, 43, 2001
|
|
1U7T
 
 | | Crystal Structure of ABAD/HSD10 with a Bound Inhibitor | | Descriptor: | 1-AZEPAN-1-YL-2-PHENYL-2-(4-THIOXO-1,4-DIHYDRO-PYRAZOLO[3,4-D]PYRIMIDIN-5-YL)ETHANONE ADDUCT, 3-hydroxyacyl-CoA dehydrogenase type II, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kissinger, C.R, Rejto, P.A, Pelletier, L.A, Showalter, R.E, Villafranca, J.E. | | Deposit date: | 2004-08-04 | | Release date: | 2004-10-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of human ABAD/HSD10 with a bound inhibitor: implications for design of Alzheimer's disease therapeutics
J.Mol.Biol., 342, 2004
|
|
1CW1
 
 | |
2RDT
 
 | | Crystal Structure of Human Glycolate Oxidase (GO) in Complex with CDST | | Descriptor: | 5-(dodecylthio)-1H-1,2,3-triazole-4-carboxylic acid, FLAVIN MONONUCLEOTIDE, Hydroxyacid oxidase 1 | | Authors: | Murray, M.S, Holmes, R.P, Lowther, W.T. | | Deposit date: | 2007-09-25 | | Release date: | 2008-02-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Active Site and Loop 4 Movements within Human Glycolate Oxidase: Implications for Substrate Specificity and Drug Design.
Biochemistry, 47, 2008
|
|
3P7Y
 
 | |
3CD7
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[5-(ANILINOCARBONYL)-3,4-BIS(4-FLUOROPHENYL)-1-ISOPROPYL-1H-PYRROL-2-YL]-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
1DLJ
 
 | | THE FIRST STRUCTURE OF UDP-GLUCOSE DEHYDROGENASE (UDPGDH) REVEALS THE CATALYTIC RESIDUES NECESSARY FOR THE TWO-FOLD OXIDATION | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Campbell, R.E, Mosimann, S.C, van de Rijn, I, Tanner, M.E, Strynadka, N.C.J. | | Deposit date: | 1999-12-09 | | Release date: | 2000-05-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The first structure of UDP-glucose dehydrogenase reveals the catalytic residues necessary for the two-fold oxidation.
Biochemistry, 39, 2000
|
|
5GLY
 
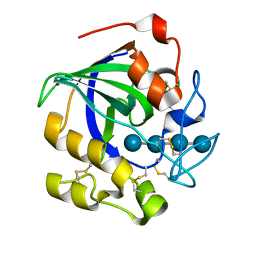 | | Crystal structure of a glycoside hydrolase in complex with cellotetrose from Thielavia terrestris NRRL 8126 | | Descriptor: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
4J1T
 
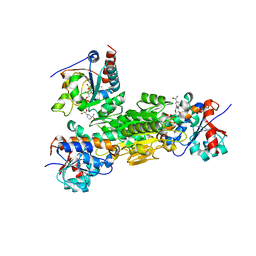 | | Crystal structure of Thermus thermophilus transhydrogenase heterotrimeric complex of the Alpha1 subunit dimer with the NADP binding domain (domain III) of the Beta subunit in P2(1) | | Descriptor: | GLYCEROL, NAD(P) transhydrogenase subunit beta, NAD/NADP transhydrogenase alpha subunit 1, ... | | Authors: | Yamaguchi, M, Leung, J, Schurig Briccio, L.A, Gennis, R.B, Stout, C.D. | | Deposit date: | 2013-02-02 | | Release date: | 2014-02-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure analysis of Thermus thermophilus transhydrogenase soluble domains
To be Published
|
|
3P81
 
 | |
4BB3
 
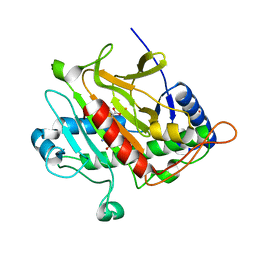 | | Isopenicillin N synthase with the dipeptide substrate analogue AhC | | Descriptor: | (2S)-2-azanyl-6-oxidanylidene-6-[[(2S)-1-oxidanyl-1-oxidanylidene-4-sulfanyl-butan-2-yl]amino]hexanoic acid, FE (III) ION, ISOPENICILLIN N SYNTHASE, ... | | Authors: | Daruzzaman, A, Clifton, I.J, Rutledge, P.J. | | Deposit date: | 2012-09-19 | | Release date: | 2013-04-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of Isopenicillin N Synthase with a Dipeptide Substrate Analogue.
Arch.Biochem.Biophys., 530, 2013
|
|
2RBW
 
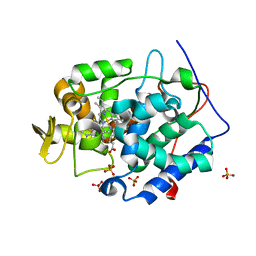 | | Cytochrome C Peroxidase W191G in complex with 1,2-dimethyl-1h-pyridin-5-amine | | Descriptor: | 5-amino-1,2-dimethylpyridinium, Cytochrome C Peroxidase, PHOSPHATE ION, ... | | Authors: | Graves, A.P, Boyce, S.E, Shoichet, B.K. | | Deposit date: | 2007-09-19 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rescoring docking hit lists for model cavity sites: predictions and experimental testing.
J.Mol.Biol., 377, 2008
|
|
6AG0
 
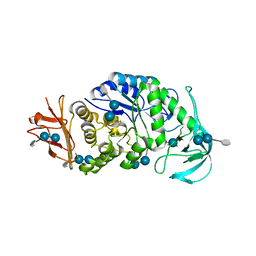 | | The X-ray Crystallographic Structure of Maltooligosaccharide-forming Amylase from Bacillus stearothermophilus STB04 | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-amylase, CALCIUM ION | | Authors: | Li, Z.F, Li, Y.L, Ban, X.F, Zhang, C.Y, Jin, T.C, Xie, X.F, Gu, Z.B, Li, C.M. | | Deposit date: | 2018-08-09 | | Release date: | 2018-10-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a maltooligosaccharide-forming amylase from Bacillus stearothermophilus STB04.
Int.J.Biol.Macromol., 138, 2019
|
|
3PYX
 
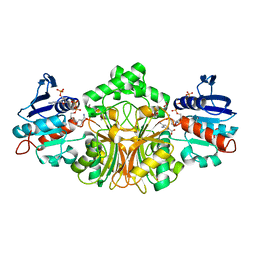 | | Crystals Structure of Aspartate beta-Semialdehyde Dehydrogenase complex with NADP and 2-aminoterephthalate | | Descriptor: | 1,2-ETHANEDIOL, 2-aminobenzene-1,4-dicarboxylic acid, Aspartate-semialdehyde dehydrogenase, ... | | Authors: | Pavlovsky, A.G, Viola, R.E. | | Deposit date: | 2010-12-13 | | Release date: | 2012-01-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Characterization of Inhibitors with Selectivity against Members of a Homologous Enzyme Family.
Chem.Biol.Drug Des., 79, 2012
|
|
4JWU
 
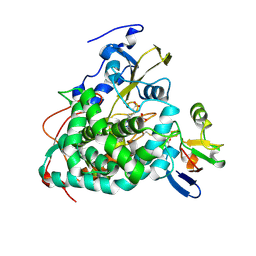 | | Crystal structure of Cytochrome P450cam-putidaredoxin complex | | Descriptor: | 1,1'-hexane-1,6-diyldipyrrolidine-2,5-dione, CALCIUM ION, Camphor 5-monooxygenase, ... | | Authors: | Tripathi, S.M, Li, H, Poulos, T.L. | | Deposit date: | 2013-03-27 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for effector control and redox partner recognition in cytochrome P450.
Science, 340, 2013
|
|
3L07
 
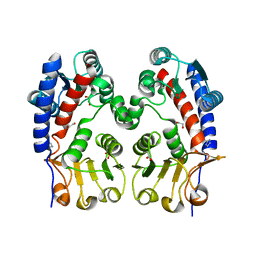 | | Methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase, putative bifunctional protein folD from Francisella tularensis. | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Bifunctional protein folD, ... | | Authors: | Osipiuk, J, Maltseva, N, Mulligan, R, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-09 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | X-ray crystal structure of methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate cyclohydrolase, putative bifunctional protein folD from Francisella tularensis.
To be Published
|
|
3CV6
 
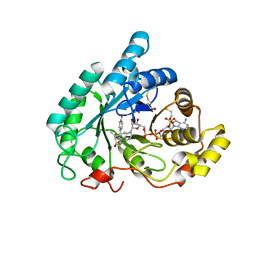 | | The crystal structure of mouse 17-alpha hydroxysteroid dehydrogenase GG225.226PP mutant in complex with inhibitor and cofactor NADP+. | | Descriptor: | 4-[(1R,2S)-1-ethyl-2-(4-hydroxyphenyl)butyl]phenol, Aldo-keto reductase family 1 member C21, BETA-MERCAPTOETHANOL, ... | | Authors: | Dhagat, U, El-Kabbani, O. | | Deposit date: | 2008-04-17 | | Release date: | 2009-03-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the G225P/G226P mutant of mouse 3(17)alpha-hydroxysteroid dehydrogenase (AKR1C21) ternary complex: implications for the binding of inhibitor and substrate.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2DLD
 
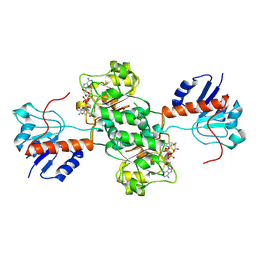 | | D-LACTATE DEHYDROGENASE COMPLEXED WITH NADH AND OXAMATE | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, D-LACTATE DEHYDROGENASE, OXAMIC ACID | | Authors: | Dunn, C.R, Holbrook, J.J. | | Deposit date: | 1995-10-28 | | Release date: | 1996-03-14 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Dehydrogenases Engineering to Correct Substrate Inhibition in a Commercial Dehydrogenase
To be Published
|
|
2N3D
 
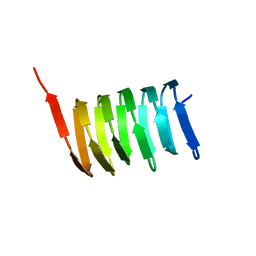 | | Atomic structure of the cytoskeletal bactofilin BacA revealed by solid-state NMR | | Descriptor: | Bactofilin A | | Authors: | Shi, C, Fricke, P, Lin, L, Chevelkov, V, Wegstroth, M, Giller, K, Becker, S, Thanbichler, M, Lange, A. | | Deposit date: | 2015-05-29 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Atomic-resolution structure of cytoskeletal bactofilin by solid-state NMR.
Sci Adv, 1, 2015
|
|
