8DSY
 
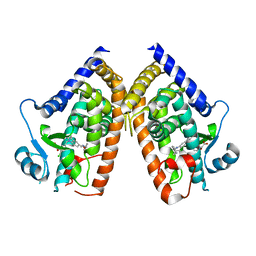 | | PPARg bound to inverse agonist H3B-343 | | Descriptor: | Peroxisome proliferator-activated receptor gamma, {5-[(5-{[(4-tert-butylphenyl)methyl]carbamoyl}-2,3-dimethyl-1H-indol-1-yl)methyl]-2-chlorophenoxy}acetic acid | | Authors: | Larsen, N.A. | | Deposit date: | 2022-07-24 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Biochemical and structural basis for the pharmacological inhibition of nuclear hormone receptor PPAR gamma by inverse agonists.
J.Biol.Chem., 298, 2022
|
|
2AXY
 
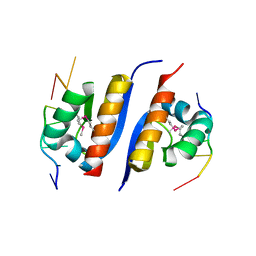 | | Crystal Structure of KH1 domain of human Poly(C)-binding protein-2 with C-rich strand of human telomeric DNA | | Descriptor: | C-rich strand of human telomeric dna, Poly(rC)-binding protein 2 | | Authors: | Du, Z, Lee, J.K, Tjhen, R.J, Li, S, Stroud, R.M, James, T.L. | | Deposit date: | 2005-09-06 | | Release date: | 2005-09-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the First KH Domain of Human Poly(C)-binding Protein-2 in Complex with a C-rich Strand of Human Telomeric DNA at 1.7 A
J.Biol.Chem., 280, 2005
|
|
6QMC
 
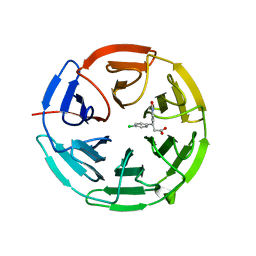 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{S})-3-(4-chlorophenyl)-3-(2-oxidanylidene-1~{H}-pyridin-4-yl)propanoic acid, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
7RDN
 
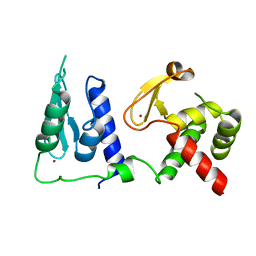 | | Crystal structure of S. cerevisiae pre-mRNA leakage protein 39 (Pml39) | | Descriptor: | Pre-mRNA leakage protein 39, ZINC ION | | Authors: | Hashimoto, H, Ramirez, D.H, Pawlak, N, Blobel, G, Palancade, B, Debler, E.W. | | Deposit date: | 2021-07-09 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure of the pre-mRNA leakage 39-kDa protein reveals a single domain of integrated zf-C3HC and Rsm1 modules.
Sci Rep, 12, 2022
|
|
4ZG0
 
 | | Crystal structure of Mouse Syndesmos protein | | Descriptor: | Protein syndesmos | | Authors: | Lee, I, Kim, H, Yoo, J, Cho, H, Lee, W. | | Deposit date: | 2015-04-22 | | Release date: | 2016-04-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.006 Å) | | Cite: | Crystal structure of syndesmos and its interaction with Syndecan-4 proteoglycan
Biochem.Biophys.Res.Commun., 463, 2015
|
|
1GV0
 
 | | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases | | Descriptor: | MALATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Dalhus, B, Sarinen, M, Sauer, U.H, Eklund, P, Johansson, K, Karlsson, A, Ramaswamy, S, Bjork, A, Synstad, B, Naterstad, K, Sirevag, R, Eklund, H. | | Deposit date: | 2002-02-04 | | Release date: | 2002-02-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for Thermophilic Protein Stability: Structures of Thermophilic and Mesophilic Malate Dehydrogenases
J.Mol.Biol., 318, 2002
|
|
8F28
 
 | | Lysozyme Structures from Single-Entity Crystallization Method NanoAC | | Descriptor: | ACETATE ION, CHLORIDE ION, Lysozyme C, ... | | Authors: | Yang, R, Sankaran, B, Ogbonna, E, Wang, G. | | Deposit date: | 2022-11-07 | | Release date: | 2023-07-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | A Single-Entity Method for Actively Controlled Nucleation and High-Quality Protein Crystal Synthesis.
Anal.Chem., 95, 2023
|
|
7UDI
 
 | |
7UQO
 
 | | Pathogenesis related 10-10 (S)-tetrahydropapaverine complex | | Descriptor: | (1~{S})-1-[(3,4-dimethoxyphenyl)methyl]-6,7-dimethoxy-1,2,3,4-tetrahydroisoquinoline, Pathogenesis Related 10-10 protein | | Authors: | Carr, S.C, Ng, K.K.S. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Alkaloid binding to opium poppy major latex proteins triggers structural modification and functional aggregation.
Nat Commun, 13, 2022
|
|
7UQN
 
 | | Pathogenesis related 10-10 noscapine complex | | Descriptor: | Pathogenesis Related 10-10 protein, noscapine | | Authors: | Carr, S.C, Ng, K.K.S. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Alkaloid binding to opium poppy major latex proteins triggers structural modification and functional aggregation.
Nat Commun, 13, 2022
|
|
6QMJ
 
 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{S})-3-(7-methoxy-1-methyl-benzotriazol-5-yl)-3-[4-methyl-3-[[methyl(phenylsulfonyl)amino]methyl]phenyl]propanoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
7UQL
 
 | | Pathogenesis related 10-10 app from | | Descriptor: | Pathogenesis Related 10-10 protein | | Authors: | Carr, S.C, Ng, K.K.S. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Alkaloid binding to opium poppy major latex proteins triggers structural modification and functional aggregation.
Nat Commun, 13, 2022
|
|
7UQM
 
 | | Pathogenesis related 10-10 papaverine complex | | Descriptor: | 1-(3,4-DIMETHOXYBENZYL)-6,7-DIMETHOXYISOQUINOLINE, Pathogenesis Related 10-10 protein | | Authors: | Carr, S.C, Ng, K.K.S. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Alkaloid binding to opium poppy major latex proteins triggers structural modification and functional aggregation.
Nat Commun, 13, 2022
|
|
4TWU
 
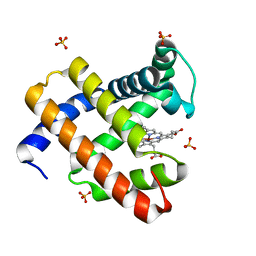 | |
8EO2
 
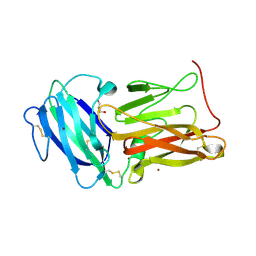 | | Lufaxin a bifunctional inhibitor of complement and coagulation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BROMIDE ION, ... | | Authors: | Andersen, J.F, Strayer, E.C. | | Deposit date: | 2022-10-01 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | A bispecific inhibitor of complement and coagulation blocks activation in complementopathy models via a novel mechanism.
Blood, 141, 2023
|
|
8ENU
 
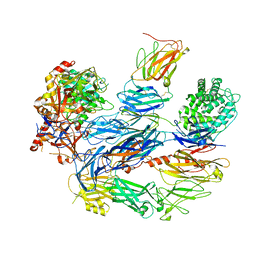 | | Structure of the C3bB proconvertase in complex with lufaxin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Complement C3 beta chain, Complement C3b alpha' chain, ... | | Authors: | Andersen, J.F, Lei, H. | | Deposit date: | 2022-09-30 | | Release date: | 2023-08-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | A bispecific inhibitor of complement and coagulation blocks activation in complementopathy models via a novel mechanism.
Blood, 141, 2023
|
|
4TUG
 
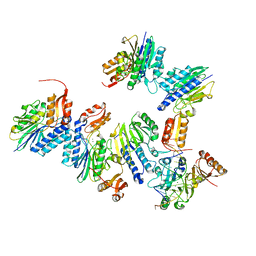 | | Crystal structure of MjMre11-DNA2 complex | | Descriptor: | DNA (5'-D(P*CP*TP*GP*TP*CP*CP*TP*AP*CP*GP*TP*GP*CP*CP*A)-3'), DNA (5'-D(P*GP*CP*AP*CP*GP*TP*AP*GP*GP*AP*CP*AP*GP*C)-3'), DNA double-strand break repair protein Mre11, ... | | Authors: | Sung, S, Cho, Y. | | Deposit date: | 2014-06-24 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | DNA end recognition by the Mre11 nuclease dimer: insights into resection and repair of damaged DNA.
Embo J., 33, 2014
|
|
4TWV
 
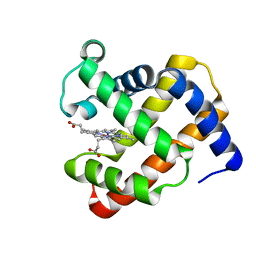 | |
7YKD
 
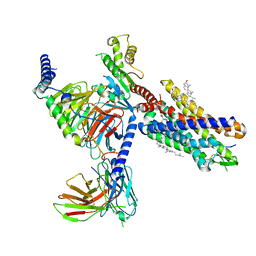 | | Cryo-EM structure of the human chemerin receptor 1 complex with the C-terminal nonapeptide of chemerin | | Descriptor: | CHOLESTEROL, Chemerin-like receptor 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, G, Liao, Q, Ye, R.D, Wang, J. | | Deposit date: | 2022-07-22 | | Release date: | 2023-04-19 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Cryo-EM structure of the human chemerin receptor 1-Gi protein complex bound to the C-terminal nonapeptide of chemerin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4X3V
 
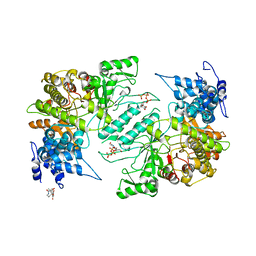 | | Crystal structure of human ribonucleotide reductase 1 bound to inhibitor | | Descriptor: | N~6~-{N-[(1,3-dioxo-1,3-dihydro-2H-isoindol-2-yl)acetyl]-2-methyl-D-alanyl}-D-lysine, Ribonucleoside-diphosphate reductase large subunit, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Dealwis, C.G, Ahmad, M.F, Alam, I. | | Deposit date: | 2014-12-01 | | Release date: | 2015-11-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Identification of Non-nucleoside Human Ribonucleotide Reductase Modulators.
J.Med.Chem., 58, 2015
|
|
6QV7
 
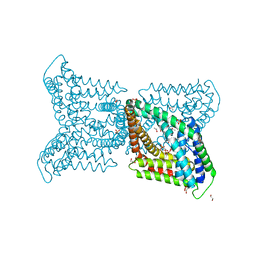 | |
6QVA
 
 | |
7R2E
 
 | | Structure of human Senp7 with SUMO2 | | Descriptor: | Sentrin-specific protease 7, Small ubiquitin-related modifier 3, prop-2-en-1-amine | | Authors: | Reverter, D, Li, Y. | | Deposit date: | 2022-02-04 | | Release date: | 2022-12-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Basis for the SUMO2 Isoform Specificity of SENP7.
J.Mol.Biol., 434, 2022
|
|
7YK3
 
 | | Crystal structure of DarTG toxin-antitoxin complex from Mycobacterium tuberculosis | | Descriptor: | DNA ADP-ribosyl glycohydrolase, DNA ADP-ribosyl transferase, PHOSPHATE ION | | Authors: | Deep, A, Kaur, J, Singh, L, Thakur, K.G. | | Deposit date: | 2022-07-21 | | Release date: | 2023-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into DarT toxin neutralization by cognate DarG antitoxin: ssDNA mimicry by DarG C-terminal domain keeps the DarT toxin inhibited.
Structure, 31, 2023
|
|
7RDG
 
 | |
