1NAS
 
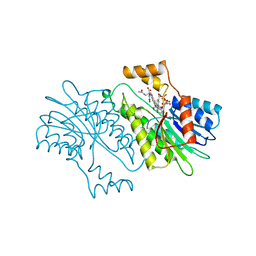 | | SEPIAPTERIN REDUCTASE COMPLEXED WITH N-ACETYL SEROTONIN | | Descriptor: | N-ACETYL SEROTONIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, OXALOACETATE ION, ... | | Authors: | Auerbach, G, Herrmann, A, Bacher, A, Huber, R. | | Deposit date: | 1998-03-26 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The 1.25 A crystal structure of sepiapterin reductase reveals its binding mode to pterins and brain neurotransmitters.
EMBO J., 16, 1997
|
|
3BUR
 
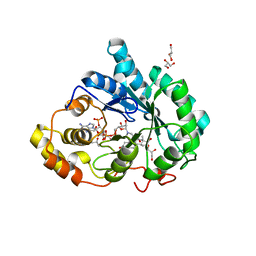 | |
3T3Q
 
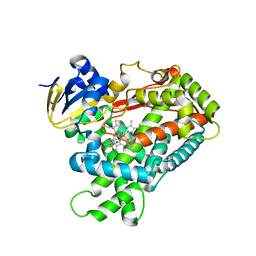 | | Human Cytochrome P450 2A6 I208S/I300F/G301A/S369G in complex with Pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
1JZI
 
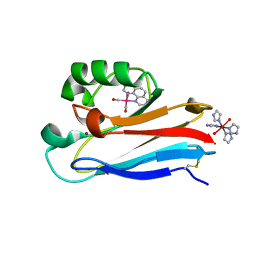 | | Pseudomonas aeruginosa Azurin Re(phen)(CO)3(His83) | | Descriptor: | (1,10 PHENANTHROLINE)-(TRI-CARBON MONOXIDE) RHENIUM (I), AZURIN, COPPER (II) ION, ... | | Authors: | Crane, B.R, Di Bilio, A.J, Winkler, J.R, Gray, H.B. | | Deposit date: | 2001-09-16 | | Release date: | 2001-10-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Electron tunneling in single crystals of Pseudomonas aeruginosa azurins.
J.Am.Chem.Soc., 123, 2001
|
|
3WLF
 
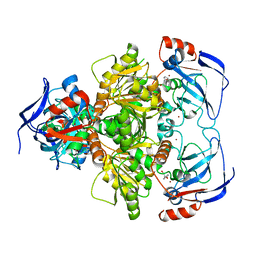 | | Crystal structure of (R)-carbonyl reductase from Candida Parapsilosis in complex with (R)-1-phenyl-1,2-ethanediol | | Descriptor: | (1R)-1-phenylethane-1,2-diol, (R)-specific carbonyl reductase, ZINC ION | | Authors: | Wang, S.S, Nie, Y, Xu, Y, Zhang, R.Z, Huang, C.H, Chan, H.C, Guo, R.T, Xiao, R. | | Deposit date: | 2013-11-09 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Unconserved substrate-binding sites direct the stereoselectivity of medium-chain alcohol dehydrogenase
Chem.Commun.(Camb.), 50, 2014
|
|
6JVC
 
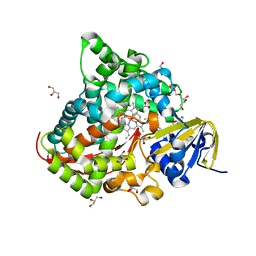 | | Structure of the Cobalt Protoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional cytochrome P450/NADPH--P450 reductase, ... | | Authors: | Stanfield, J.K, Matsumoto, A, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-04-16 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
3KRK
 
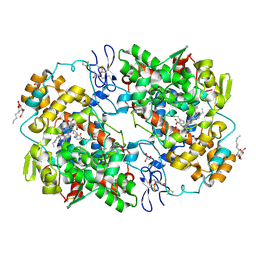 | | X-ray crystal structure of arachidonic acid bound in the cyclooxygenase channel of L531F murine COX-2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACRYLIC ACID, ... | | Authors: | Vecchio, A.J, Simmons, D.M, Malkowski, M.G. | | Deposit date: | 2009-11-18 | | Release date: | 2010-05-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of fatty acid substrate binding to cyclooxygenase-2.
J.Biol.Chem., 285, 2010
|
|
5DKE
 
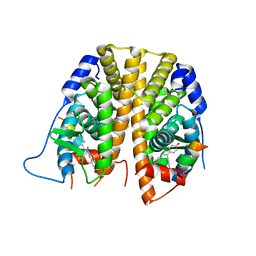 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 3-naphthyl-substituted, methyl, cis-diaryl-ethylene compound 4,4'-[2-(naphthalen-2-yl)prop-1-ene-1,1-diyl]diphenol | | Descriptor: | 4,4'-[2-(naphthalen-2-yl)prop-1-ene-1,1-diyl]diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-03 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
5DL4
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a phenylamino-substituted, methyl, triaryl-ethylene derivative 4,4'-{2-[3-(phenylamino)phenyl]prop-1-ene-1,1-diyl}diphenol | | Descriptor: | 4,4'-{2-[3-(phenylamino)phenyl]prop-1-ene-1,1-diyl}diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
3FX4
 
 | | Porcine aldehyde reductase in ternary complex with inhibitor | | Descriptor: | Alcohol dehydrogenase [NADP+], NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, ... | | Authors: | Carbone, V, El-Kabbani, O. | | Deposit date: | 2009-01-20 | | Release date: | 2009-12-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of aldehyde reductase in ternary complex with a 5-arylidene-2,4-thiazolidinedione aldose reductase inhibitor.
Eur.J.Med.Chem., 45, 2010
|
|
3T3R
 
 | | Human Cytochrome P450 2A6 in complex with Pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
4Q3C
 
 | | PylD cocrystallized with L-Lysine-Ne-L-lysine and NAD+ | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Quitterer, F, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2014-04-11 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Formation of Pyrroline and Tetrahydropyridine Rings in Amino Acids Catalyzed by Pyrrolysine Synthase (PylD).
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4I08
 
 | | Crystal structure of beta-ketoacyl-acyl carrier protein reductase (FabG) from Vibrio cholerae in complex with NADPH | | Descriptor: | 1,2-ETHANEDIOL, 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hou, J, Chruszcz, M, Zheng, H, Osinski, T, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-16 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.063 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
4I62
 
 | | 1.05 Angstrom crystal structure of an amino acid ABC transporter substrate-binding protein AbpA from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine | | Descriptor: | ARGININE, Amino acid ABC transporter, periplasmic amino acid-binding protein, ... | | Authors: | Stogios, P.J, Kudritska, M, Wawrzak, Z, Minasov, G, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-11-29 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 1.05 Angstrom crystal structure of an amino acid ABC transporter substrate-binding protein from Streptococcus pneumoniae Canada MDR_19A bound to L-arginine
To be Published
|
|
6K61
 
 | | Cryo-EM structure of the tetrameric photosystem I from a heterocyst-forming cyanobacterium Anabaena sp. PCC7120 | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Zheng, L, Li, Y, Li, X, Zhong, Q, Li, N, Zhang, K, Zhang, Y, Chu, H, Ma, C, Li, G, Zhao, J, Gao, N. | | Deposit date: | 2019-05-31 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | Structural and functional insights into the tetrameric photosystem I from heterocyst-forming cyanobacteria.
Nat.Plants, 5, 2019
|
|
4Q3A
 
 | | PylD cocrystallized with L-Lysine-Ne-3S-methyl-L-ornithine and NAD+ | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Quitterer, F, Beck, P, Bacher, A, Groll, M. | | Deposit date: | 2014-04-11 | | Release date: | 2014-04-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Formation of Pyrroline and Tetrahydropyridine Rings in Amino Acids Catalyzed by Pyrrolysine Synthase (PylD).
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
6A36
 
 | | Mandelate oxidase mutant-Y128F with the 3-fluoropyruvic acid FMN adduct | | Descriptor: | 1-deoxy-1-{5-[(1S)-2-fluoro-1-hydroxyethyl]-7,8-dimethyl-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl}-5-O-phosphono-D-ribitol, 1-{5-[(3S)-3-carboxy-4-fluoro-3-hydroxybutanoyl]-7,8-dimethyl-2,4-dioxo-1,3,4,5-tetrahydrobenzo[g]pteridin-10(2H)-yl}-1-deoxy-5-O-phosphono-D-ribitol, 4-hydroxymandelate oxidase | | Authors: | Li, T.L, Lin, K.H. | | Deposit date: | 2018-06-15 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural and chemical trapping of flavin-oxide intermediates reveals substrate-directed reaction multiplicity.
Protein Sci., 29, 2020
|
|
1WVE
 
 | | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon its Binding to the Cytochrome Subunit | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-cresol dehydrogenase [hydroxylating] cytochrome c subunit, 4-cresol dehydrogenase [hydroxylating] flavoprotein subunit, ... | | Authors: | Cunane, L.M, Chen, Z.-W, McIntire, W.S, Mathews, F.S. | | Deposit date: | 2004-12-15 | | Release date: | 2005-03-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | p-Cresol Methylhydroxylase: Alteration of the Structure of the Flavoprotein Subunit upon Its Binding to the Cytochrome Subunit
Biochemistry, 44, 2005
|
|
3CD7
 
 | | Thermodynamic and structure guided design of statin hmg-coa reductase inhibitors | | Descriptor: | (3R,5R)-7-[5-(ANILINOCARBONYL)-3,4-BIS(4-FLUOROPHENYL)-1-ISOPROPYL-1H-PYRROL-2-YL]-3,5-DIHYDROXYHEPTANOIC ACID, 3-hydroxy-3-methylglutaryl-coenzyme A reductase | | Authors: | Pavlovsky, A, Sarver, R.W, Harris, M.S, Finzel, B.C. | | Deposit date: | 2008-02-26 | | Release date: | 2008-06-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Thermodynamic and structure guided design of statin based inhibitors of 3-hydroxy-3-methylglutaryl coenzyme a reductase.
J.Med.Chem., 51, 2008
|
|
5DMF
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in complex with a 4-fluorophenylamino-substituted, methyl triaryl-ethylene derivative 4,4'-(2-{3-[(4-fluorophenyl)amino]phenyl}prop-1-ene-1,1-diyl)diphenol | | Descriptor: | 4,4'-(2-{3-[(4-fluorophenyl)amino]phenyl}prop-1-ene-1,1-diyl)diphenol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-08 | | Release date: | 2016-05-04 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
2RC2
 
 | |
4DGQ
 
 | | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia | | Descriptor: | 1,2-ETHANEDIOL, Non-heme chloroperoxidase | | Authors: | Gardberg, A.S, Edwards, T.E, Abendroth, J.A, Staker, B, Stewart, L, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2012-01-26 | | Release date: | 2012-03-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of Non-heme chloroperoxidase from Burkholderia cenocepacia
To be Published
|
|
4DYY
 
 | | Crystal Structure of the Cu-adduct of Human H-Ferritin variant MIC1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Tezcan, F.A, Huard, D.J.E. | | Deposit date: | 2012-02-29 | | Release date: | 2013-01-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Re-engineering protein interfaces yields copper-inducible ferritin cage assembly.
Nat.Chem.Biol., 9, 2013
|
|
2RAA
 
 | |
2HUO
 
 | | Crystal structure of mouse myo-inositol oxygenase in complex with substrate | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, FE (III) ION, FORMIC ACID, ... | | Authors: | Brown, P.M, Caradoc-Davies, T.T, Dickson, J.M.J, Cooper, G.J.S, Loomes, K.M, Baker, E.N. | | Deposit date: | 2006-07-27 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a substrate complex of myo-inositol oxygenase, a di-iron oxygenase with a key role in inositol metabolism.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
