6CC0
 
 | |
5J4L
 
 | | Apo-structure of humanised RadA-mutant humRadA22F | | Descriptor: | CHLORIDE ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-01 | | Release date: | 2016-10-19 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JEE
 
 | | Apo-structure of humanised RadA-mutant humRadA26F | | Descriptor: | CALCIUM ION, DNA repair and recombination protein RadA | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-18 | | Release date: | 2016-10-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Engineering Archeal Surrogate Systems for the Development of Protein-Protein Interaction Inhibitors against Human RAD51.
J.Mol.Biol., 428, 2016
|
|
5JFG
 
 | | Structure of humanised RadA-mutant humRadA22F in complex with peptide FHTA | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, DNA repair and recombination protein RadA, ... | | Authors: | Fischer, G, Marsh, M, Moschetti, T, Sharpe, T, Scott, D, Morgan, M, Ng, H, Skidmore, J, Venkitaraman, A, Abell, C, Blundell, T.L, Hyvonen, M. | | Deposit date: | 2016-04-19 | | Release date: | 2016-10-26 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Humanisation of RadA from Pyrococcus furiosus
To Be Published
|
|
2EXS
 
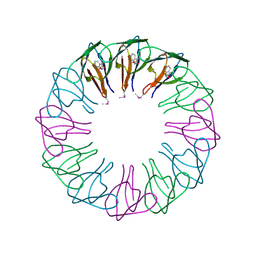 | | TRAP3 (engineered TRAP) | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB | | Authors: | Heddle, J.G, Yokoyama, T, Yamashita, I, Park, S.Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Rounding up: Engineering 12-Membered Rings from the Cyclic 11-Mer TRAP
Structure, 14, 2006
|
|
3HL8
 
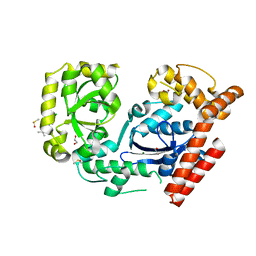 | | Crystal structure of exonuclease I in complex with inhibitor BCBP | | Descriptor: | (5R)-3-tert-butyl-1-(6-chloro-1,3-benzothiazol-2-yl)-4,5-dihydro-1H-pyrazol-5-ol, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Satyshur, K.A. | | Deposit date: | 2009-05-27 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Small-molecule tools for dissecting the roles of SSB/protein interactions in genome maintenance
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4QYG
 
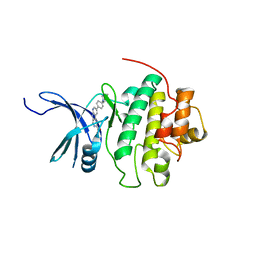 | | CHK1 kinase domain in complex with diazacarbazole compound 14 | | Descriptor: | 3-[4-(4-methylpiperazin-1-yl)phenyl]-9H-pyrrolo[2,3-b:5,4-c']dipyridine-6-carboxylic acid, Serine/threonine-protein kinase Chk1 | | Authors: | Wiesmann, C, Wu, P. | | Deposit date: | 2014-07-24 | | Release date: | 2014-12-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of the 1,7-diazacarbazole class of inhibitors of checkpoint kinase 1.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
2EXT
 
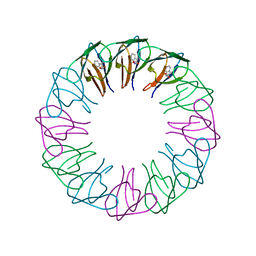 | | TRAP4 (engineered TRAP) | | Descriptor: | TRYPTOPHAN, Transcription attenuation protein mtrB | | Authors: | Heddle, J.G, Yokoyama, T, Yamashita, I, Park, S.Y, Tame, J.R.H. | | Deposit date: | 2005-11-08 | | Release date: | 2006-08-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Rounding up: Engineering 12-Membered Rings from the Cyclic 11-Mer TRAP
Structure, 14, 2006
|
|
4GCV
 
 | |
4JBZ
 
 | | Structure of Mcm10 coiled-coil region | | Descriptor: | MALTOSE-BINDING PERIPLASMIC PROTEIN FUSED WITH XENOPUS LAEVIS MCM10 COILED-COIL REGION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Du, W, Adhikary, S, Eichman, B.F. | | Deposit date: | 2013-02-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mcm10 self-association is mediated by an N-terminal coiled-coil domain.
Plos One, 8, 2013
|
|
1MXJ
 
 | | NMR solution structure of benz[a]anthracene-dG in ras codon 12,2; GGCAGXTGGTG | | Descriptor: | 1S,2R,3S,4R-TETRAHYDRO-BENZO[A]ANTHRACENE-2,3,4-TRIOL, 5'-D(*CP*AP*CP*CP*AP*CP*CP*TP*GP*CP*C)-3', 5'-D(*GP*GP*CP*AP*GP*GP*TP*GP*GP*TP*G)-3' | | Authors: | Kim, H.-Y.H, Wilkinson, A.S, Harris, C.M, Harris, T.M, Stone, M.P. | | Deposit date: | 2002-10-02 | | Release date: | 2003-03-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Minor Groove Orientation for the (1S,2R,3S,4R)-N2-[1-(1,2,3,4-tetrahydro-2,3,4-trihydroxy-benz[a]anthracenyl)]-2'-deoxyguanosyl Adduct in the N-ras Codon 12 sequence
Biochemistry, 42, 2003
|
|
4ZHN
 
 | | Crystal Structure of AlkB T208A mutant protein in complex with Co(II), 2-oxoglutarate, and methylated trinucleotide T-meA-T | | Descriptor: | 2-OXOGLUTARIC ACID, Alpha-ketoglutarate-dependent dioxygenase AlkB, COBALT (II) ION, ... | | Authors: | Ergel, B, Yu, B, Vorobiev, S.M, Forouhar, F, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2015-04-25 | | Release date: | 2015-12-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.333 Å) | | Cite: | DFT Studies of the Rate-Limiting Step in the Reaction Cycle of the Fe/2OG Dioxygenase AlkB and Related Experimental Studies
To be published
|
|
4CDN
 
 | | Crystal structure of M. mazei photolyase with its in vivo reconstituted 8-HDF antenna chromophore | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, DEOXYRIBODIPYRIMIDINE PHOTOLYASE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kiontke, S, Batschauer, A, Essen, L.-O. | | Deposit date: | 2013-11-01 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Evolutionary Aspects of Antenna Chromophore Usage by Class II Photolyases.
J.Biol.Chem., 289, 2014
|
|
4Z58
 
 | | HipB-O3 20mer complex | | Descriptor: | Antitoxin HipB, DNA (5'-D(*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*TP*AP*CP*GP*GP*GP*AP*TP*AP*A)-3') | | Authors: | Min, J, Brennan, R.G, Schumacher, M.A. | | Deposit date: | 2015-04-02 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular mechanism on hipBA gene regulation.
To be published
|
|
8CEZ
 
 | |
2YPY
 
 | |
6J8W
 
 | | Structure of MOEN5-SSO7D fusion protein in complex with lig 1 | | Descriptor: | (2S)-3-dimethoxyphosphoryloxy-2-[(2E,6E)-3,7,11-trimethyldodeca-2,6,10-trienoxy]propanoic acid, MoeN5,DNA-binding protein 7d | | Authors: | Ko, T.P, Zhang, L.L, Chen, C.C, Guo, R.T. | | Deposit date: | 2019-01-21 | | Release date: | 2019-04-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Complex structures of MoeN5 with substrate analogues suggest sequential catalytic mechanism.
Biochem. Biophys. Res. Commun., 511, 2019
|
|
4HV0
 
 | | Structure and Function of AvtR, a Novel Transcriptional Regulator from a Hyperthermophilic Archaeal Lipothrixvirus | | Descriptor: | AvtR | | Authors: | Peixeiro, N, Keller, J, Collinet, B, Leulliot, N, Campanacci, V, Cortez, D, Cambillau, C, Nitta, K.R, Vincentelli, R, Forterre, P, Prangishvili, D, Sezonov, G, van Tilbeurgh, H. | | Deposit date: | 2012-11-05 | | Release date: | 2012-11-21 | | Last modified: | 2012-12-26 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure and Function of AvtR, a Novel Transcriptional Regulator from a Hyperthermophilic Archaeal Lipothrixvirus.
J.Virol., 87, 2013
|
|
2IYN
 
 | | The co-factor-induced pre-active conformation in PhoB | | Descriptor: | MAGNESIUM ION, PHOSPHATE REGULON TRANSCRIPTIONAL REGULATORY PROTEIN PHOB | | Authors: | Sola, M, Drew, D.L, Blanco, A.G, Gomis-Ruth, F.X, Coll, M. | | Deposit date: | 2006-07-19 | | Release date: | 2006-08-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The Cofactor-Induced Pre-Active Conformation in Phob.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
5A0W
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI E117A IN COMPLEX WITH ITS TARGET DNA AND IN THE PRESENCE OF 2MM MN | | Descriptor: | 25MER, ACETATE ION, CHLORIDE ION, ... | | Authors: | Molina, R, Besker, N, Prieto, J, Montoya, G, D'Abramo, M. | | Deposit date: | 2015-04-23 | | Release date: | 2016-05-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and Dynamical Characterization of the I- Dmo Catalytic Activity
To be Published
|
|
8J09
 
 | | Crystal structure of protein 3745 | | Descriptor: | Cell division control protein 45, DNA replication regulator SLD3 | | Authors: | Li, H, Yao, M. | | Deposit date: | 2023-04-10 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural and functional insights into a process of complex formation by 3745
To Be Published
|
|
2YQ0
 
 | |
2YPZ
 
 | |
2YQ1
 
 | |
6ZSG
 
 | | Human mitochondrial ribosome in complex with mRNA, A-site tRNA, P-site tRNA and E-site tRNA | | Descriptor: | 12S mitochondrial rRNA, 16S mitochondrial rRNA, 28S ribosomal protein S10, ... | | Authors: | Aibara, S, Singh, V, Modelska, A, Amunts, A. | | Deposit date: | 2020-07-15 | | Release date: | 2020-10-14 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis of mitochondrial translation.
Elife, 9, 2020
|
|
