5M5D
 
 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with mannose-alpha-1,3-D-glucal | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Petricevic, M, Sobala, L.F, Fernandes, P.Z, Raich, L, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2016-10-21 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Contribution of Shape and Charge to the Inhibition of a Family GH99 endo-alpha-1,2-Mannanase.
J. Am. Chem. Soc., 139, 2017
|
|
7NPH
 
 | | Crystal structure of Mycobacterium tuberculosis ArgC in complex with 5-methoxy-1,3-benzoxazole-2-carboxylic acid | | Descriptor: | 5-methoxy-1,3-benzoxazole-2-carboxylic acid, N-acetyl-gamma-glutamyl-phosphate reductase, PHOSPHATE ION | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-26 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
8WE7
 
 | | Human L-type voltage-gated calcium channel Cav1.2 in the presence of calciseptine at 3.2 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gao, S, Yao, X, Yan, N. | | Deposit date: | 2023-09-17 | | Release date: | 2023-12-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for human Ca v 1.2 inhibition by multiple drugs and the neurotoxin calciseptine.
Cell, 186, 2023
|
|
6ON4
 
 | | Crystal structure of the GntR-type sialoregulator NanR from Escherichia coli, in complex with sialic acid | | Descriptor: | HTH-type transcriptional repressor NanR, N-acetyl-beta-neuraminic acid, ZINC ION, ... | | Authors: | Horne, C.R, Panjikar, S, North, R.A, Dobson, R.C.J. | | Deposit date: | 2019-04-19 | | Release date: | 2020-07-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Control of the Escherichia coli sialoregulon by transcriptional repressor NanR.
J. Bacteriol., 195, 2013
|
|
6ONB
 
 | | Crystal Structure of the ZIG-8-RIG-5 IG1-IG1 heterodimer, monoclinic form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, NeuRonal IgCAM-5, ... | | Authors: | Cheng, S, Kurleto, J.D, Ozkan, E. | | Deposit date: | 2019-04-20 | | Release date: | 2019-05-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.696 Å) | | Cite: | Family of neural wiring receptors in bilaterians defined by phylogenetic, biochemical, and structural evidence.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7NNR
 
 | | Crystal structure of Mycobacterium tuberculosis ArgC in complex with xanthene-9-carboxylic acid | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 9~{H}-xanthene-9-carboxylic acid, N-acetyl-gamma-glutamyl-phosphate reductase | | Authors: | Gupta, P, Mendes, V, Blundell, T.L. | | Deposit date: | 2021-02-25 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A fragment-based approach to assess the ligandability of ArgB, ArgC, ArgD and ArgF in the L-arginine biosynthetic pathway of Mycobacterium tuberculosis
Comput Struct Biotechnol J, 19, 2021
|
|
9BW8
 
 | | Structure of P450Blt from Micromonospora sp. MW-13 in Complex with Fluorinated Biarylitide | | Descriptor: | Cytochrome P450-SU1, Fluorinated Biarylitide, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Hansen, M.H, Cryle, M.J, Zhao, Y. | | Deposit date: | 2024-05-21 | | Release date: | 2024-11-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Loss of fluorine during crosslinking by the biarylitide P450 Blt proceeds due to restricted peptide orientation.
Chem.Commun.(Camb.), 60, 2024
|
|
9BWH
 
 | | Crystal structure of cellulose oxidative enzyme with glycerol | | Descriptor: | COPPER (II) ION, Cellulose oxidative enzyme, GLYCEROL | | Authors: | Morais, M.A.B, Santos, C.A, Araujo, E.A, Santos, C.R, Morao, L.G, Motta, M.L, Murakami, M.T. | | Deposit date: | 2024-05-21 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | A metagenomic 'dark matter' enzyme catalyses oxidative cellulose conversion.
Nature, 639, 2025
|
|
1ZGL
 
 | | Crystal structure of 3A6 TCR bound to MBP/HLA-DR2a | | Descriptor: | HLA class II histocompatibility antigen, DR alpha chain, Myelin basic protein, ... | | Authors: | Li, Y, Huang, Y, Lue, J, Quandt, J.A, Martin, R, Mariuzza, R.A. | | Deposit date: | 2005-04-21 | | Release date: | 2005-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of a human autoimmune TCR bound to a myelin basic protein self-peptide and a multiple sclerosis-associated MHC class II molecule.
Embo J., 24, 2005
|
|
5LDH
 
 | |
9BWF
 
 | | Crystal structure of cellulose oxidative enzyme without ligand | | Descriptor: | COPPER (II) ION, Cellulose oxidative enzyme | | Authors: | Morais, M.A.B, Santos, C.A, Araujo, E.A, Santos, C.R, Morao, L.G, Motta, M.L, Murakami, M.T. | | Deposit date: | 2024-05-21 | | Release date: | 2024-12-04 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A metagenomic 'dark matter' enzyme catalyses oxidative cellulose conversion.
Nature, 639, 2025
|
|
7NNI
 
 | |
6OBN
 
 | | The crystal structure of coexpressed SDS22:PP1 complex | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE (III) ION, ... | | Authors: | Choy, M.S, Moon, T.M, Bray, J.A, Archuleta, T.L, Shi, W, Peti, W, Page, R. | | Deposit date: | 2019-03-21 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | SDS22 selectively recognizes and traps metal-deficient inactive PP1.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5M2E
 
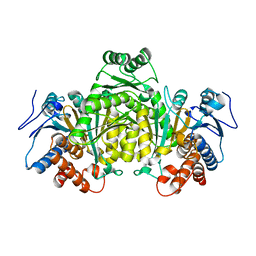 | |
6OC1
 
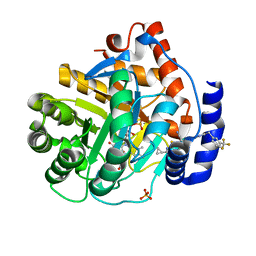 | | Crystal structure of human DHODH with TAK-632 | | Descriptor: | Dihydroorotate dehydrogenase (quinone), mitochondrial, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Durst, M.A, Lavie, A. | | Deposit date: | 2019-03-21 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Metabolic Modifier Screen Reveals Secondary Targets of Protein Kinase Inhibitors within Nucleotide Metabolism.
Cell Chem Biol, 27, 2020
|
|
6OOX
 
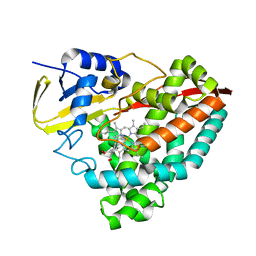 | |
8V44
 
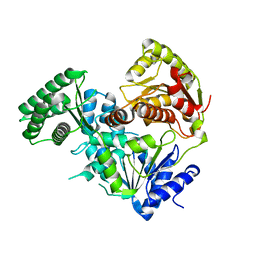 | |
5LYM
 
 | |
6OCN
 
 | |
8VJ3
 
 | |
6OHB
 
 | | E. coli Guanine Deaminase | | Descriptor: | Guanine deaminase, ZINC ION | | Authors: | Shek, R.S, French, J.B. | | Deposit date: | 2019-04-05 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Determinants for Substrate Selectivity in Guanine Deaminase Enzymes of the Amidohydrolase Superfamily.
Biochemistry, 58, 2019
|
|
5CAM
 
 | |
8VR0
 
 | |
8VR1
 
 | |
9C3H
 
 | | Structure of the CNOT3-bound human 80S ribosome with tRNA-ARG in the P-site. | | Descriptor: | 18S rRNA, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 28S rRNA, ... | | Authors: | Erzberger, J.P, Cruz, V.E. | | Deposit date: | 2024-06-01 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Specific tRNAs promote mRNA decay by recruiting the CCR4-NOT complex to translating ribosomes.
Science, 386, 2024
|
|
