5W5S
 
 | |
5VWJ
 
 | | HLA-B*58:01 presenting LTVQVARVW | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-58 alpha chain, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2017-05-22 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | HLA-B57 micropolymorphism defines the sequence and conformational breadth of the immunopeptidome.
Nat Commun, 9, 2018
|
|
5W6R
 
 | |
8Q9P
 
 | | Crystal Structure of the MADS-box/MEF2 Domain of MEF2D bound to dsDNA and HDAC5 deacetylase binding motif | | Descriptor: | HDAC5 (histone deacetylase 5) binding motif peptide: TRP-GLY-SER-GLY-GLU-VAL-LYS-LEU-ARG-LEU-GLN-GLU-PHE-LEU-LEU-SER-LYS-SER, MADS box dsDNA fw: AACTATTTATAAGA, MADS box dsNA rev:TCTTATAAATAGTT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzocato, Y, Biondi, B, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
5T6W
 
 | | HLA-B*57:01 presenting SSTRGISQLW | | Descriptor: | Beta-2-microglobulin, Decapeptide: SER-SER-THR-ARG-GLY-ILE-SER-GLN-LEU-TRP, GLYCEROL, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-01 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4Q72
 
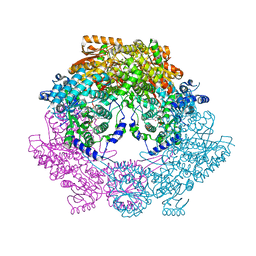 | | Crystal Structure of Bradyrhizobium japonicum Proline Utilization A (PutA) Mutant D779Y | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Proline dehydrogenase, ... | | Authors: | Tanner, J.J, Pemberton, T.A, Luo, M. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Structural Characterization of Tunnel-Perturbing Mutants in Bradyrhizobium japonicum Proline Utilization A.
Biochemistry, 53, 2014
|
|
5T70
 
 | | KIR3DL1 in complex with HLA-B*57:01 presenting TSNLQEQIGW | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Decapeptide: THR-SER-ASN-LEU-GLN-GLU-GLN-ILE-GLY-TRP, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-02 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5T6X
 
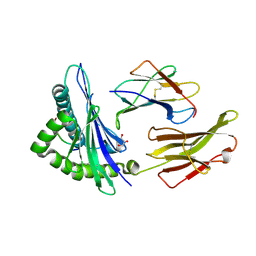 | | HLA-B*57:01 presenting TSTTSVASSW | | Descriptor: | Beta-2-microglobulin, Decapeptide: THR-SER-THR-THR-SER-VAL-ALA-SER-SER-TRP, GLYCEROL, ... | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2016-09-02 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.686 Å) | | Cite: | MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape.
Nat. Struct. Mol. Biol., 24, 2017
|
|
7JRH
 
 | |
4Q73
 
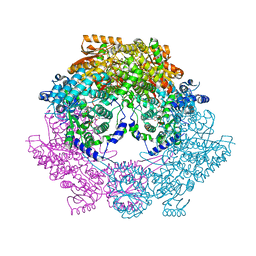 | | Crystal Structure of Bradyrhizobium japonicum Proline Utilization A (PutA) Mutant D778Y | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, Proline dehydrogenase, ... | | Authors: | Tanner, J.J, Luo, M, Pemberton, T.A. | | Deposit date: | 2014-04-23 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and Structural Characterization of Tunnel-Perturbing Mutants in Bradyrhizobium japonicum Proline Utilization A.
Biochemistry, 53, 2014
|
|
5VWD
 
 | | HLA-B*57:03 presenting LTVQVARVW | | Descriptor: | Beta-2-microglobulin, MHC class I antigen, Noamer peptide: LEU-THR-VAL-GLN-VAL-ALA-ARG-VAL-TRP | | Authors: | Pymm, P, Rossjohn, J, Vivian, J.P. | | Deposit date: | 2017-05-21 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | HLA-B57 micropolymorphism defines the sequence and conformational breadth of the immunopeptidome.
Nat Commun, 9, 2018
|
|
6FZB
 
 | | AadA in complex with ATP, magnesium and streptomycin | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kanchugal P, S, Selmer, M. | | Deposit date: | 2018-03-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase fromSalmonella enterica.
J. Biol. Chem., 293, 2018
|
|
8DDF
 
 | | Quasi-racemic mixture of L-FWF and D-FYF peptide reveals rippled beta-sheet | | Descriptor: | 1,1,1,3,3,3-hexafluoropropan-2-ol, DPN-DTY-DPN, PHE-TRP-PHE | | Authors: | Sawaya, M.R, Hazari, A, Eisenberg, D.E. | | Deposit date: | 2022-06-18 | | Release date: | 2022-09-28 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The rippled beta-sheet layer configuration-a novel supramolecular architecture based on predictions by Pauling and Corey.
Chem Sci, 13, 2022
|
|
8Q1B
 
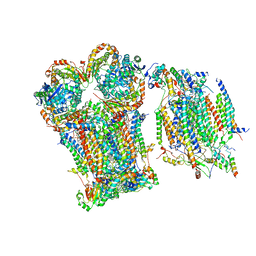 | | III2-IV1 respiratory supercomplex from S. pombe | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSHOCHOLINE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Moe, A, Brzezinski, P. | | Deposit date: | 2023-07-31 | | Release date: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure and function of the S. pombe III-IV-cyt c supercomplex.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4RTC
 
 | | Crystal structure of the green fluorescent variant, nowGFP, of the cyan Cerulean at pH 9.0 | | Descriptor: | GLYCEROL, nowGFP | | Authors: | Pletnev, V.Z, Pletneva, N.V, Pletnev, S.V. | | Deposit date: | 2014-11-14 | | Release date: | 2015-09-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of the green fluorescent protein NowGFP with an anionic tryptophan-based chromophore.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4RYS
 
 | |
8K3S
 
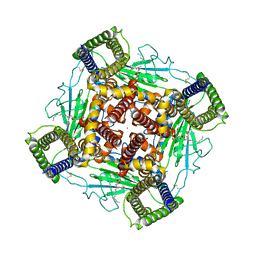 | | Structure of PKD2-F604P complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, ... | | Authors: | Chen, M.Y, Su, Q, Wang, Z.F, Yu, Y. | | Deposit date: | 2023-07-16 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Molecular and structural basis of the dual regulation of the polycystin-2 ion channel by small-molecule ligands.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
5LUH
 
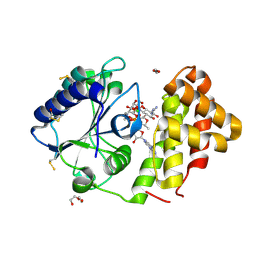 | | AadA E87Q in complex with ATP, calcium and streptomycin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Stern, A.L, Van der Verren, S.E, Selmer, M. | | Deposit date: | 2016-09-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase fromSalmonella enterica.
J.Biol.Chem., 293, 2018
|
|
5LPA
 
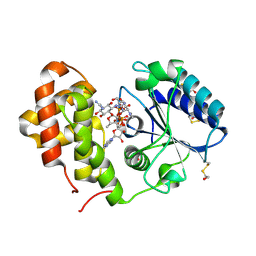 | | AadA E87Q in complex with ATP, calcium and dihydrostreptomycin | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, ... | | Authors: | Stern, A.L, Van der Verren, S.E, Selmer, M. | | Deposit date: | 2016-08-12 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase fromSalmonella enterica.
J.Biol.Chem., 293, 2018
|
|
3AM7
 
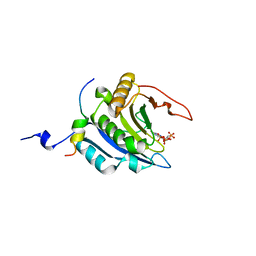 | | Crystal structure of the ternary complex of eIF4E-M7GTP-4EBP2 peptide | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, Eukaryotic translation initiation factor 4E, Eukaryotic translation initiation factor 4E-binding protein 2 | | Authors: | Tomoo, K, Fukuyo, A, In, Y, Ishida, T. | | Deposit date: | 2010-08-17 | | Release date: | 2011-08-17 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural scaffold for eIF4E binding selectivity of 4E-BP isoforms: crystal structure of eIF4E binding region of 4E-BP2 and its comparison with that of 4E-BP1.
J.Pept.Sci., 17, 2011
|
|
3NSN
 
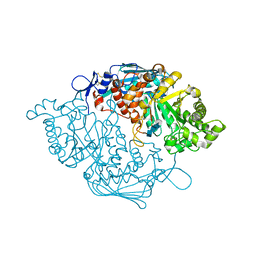 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 complexed with TMG-chitotriomycin | | Descriptor: | 2-deoxy-2-(trimethylammonio)-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
3NSM
 
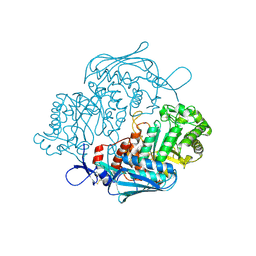 | | Crystal Structure of insect beta-N-acetyl-D-hexosaminidase OfHex1 from Ostrinia furnacalis | | Descriptor: | N-acetylglucosaminidase | | Authors: | Zhang, H, Liu, T, Liu, F, Yang, Q, Shen, X. | | Deposit date: | 2010-07-02 | | Release date: | 2010-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Determinants of an Insect {beta}-N-Acetyl-D-hexosaminidase Specialized as a Chitinolytic Enzyme
J.Biol.Chem., 286, 2011
|
|
1MPD
 
 | |
5XE4
 
 | |
1MPB
 
 | |
