6CWH
 
 | |
7WU7
 
 | | Prefoldin-tubulin-TRiC complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Prefoldin subunit 1, Prefoldin subunit 2, ... | | Authors: | Gestaut, D, Zhao, Y, Park, J, Ma, B, Leitner, A, Collier, M, Pintilie, G, Roh, S.-H, Chiu, W, Frydman, J. | | Deposit date: | 2022-02-07 | | Release date: | 2022-12-21 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural visualization of the tubulin folding pathway directed by human chaperonin TRiC/CCT.
Cell, 185, 2022
|
|
7WJU
 
 | | Cryo-EM structure of the AsCas12f1-sgRNAv1-dsDNA ternary complex | | Descriptor: | Non-target strand, OrfB_Zn_ribbon domain-containing protein, Target strand, ... | | Authors: | Wu, Z, Liu, D, Shen, H, Ji, Q. | | Deposit date: | 2022-01-07 | | Release date: | 2023-01-18 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.69 Å) | | Cite: | Structure-directed functional evolution of the miniature CRISPR-AsCas12f1 system
To Be Published
|
|
1DK2
 
 | | REFINED SOLUTION STRUCTURE OF THE N-TERMINAL DOMAIN OF DNA POLYMERASE BETA | | Descriptor: | DNA POLYMERASE BETA | | Authors: | Maciejewski, M.W, Prasad, R, Liu, D.-J, Wilson, S.H, Mullen, G.P. | | Deposit date: | 1999-12-06 | | Release date: | 2000-02-14 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Backbone dynamics and refined solution structure of the N-terminal domain of DNA polymerase beta. Correlation with DNA binding and dRP lyase activity.
J.Mol.Biol., 296, 2000
|
|
6GAZ
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the 28S ribosomal subunit. | | Descriptor: | 12S ribosomal RNA, mitochondrial, 28S ribosomal protein S18b, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
8RGG
 
 | | Structure of dynein-2 intermediate chain DYNC2I2 (WDR34) in complex with dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Dynein light chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Mladenov, M, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
3NIB
 
 | | Teg14 Apo | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, GLYCEROL, Teg14 | | Authors: | Bick, M.J, Banik, J.J, Darst, S.A, Brady, S.F. | | Deposit date: | 2010-06-15 | | Release date: | 2010-12-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The 2.7 A resolution structure of the glycopeptide sulfotransferase Teg14
Acta Crystallogr.,Sect.D, 66, 2010
|
|
8RGH
 
 | | Structure of dynein-2 intermediate chain DYNC2I1 (WDR60) in complex with the dynein-2 heavy chain DYNC2H1. | | Descriptor: | Cytoplasmic dynein 2 intermediate chain 1, Cytoplasmic dynein 2 intermediate chain 2, Cytoplasmic dynein 2 light intermediate chain 1, ... | | Authors: | Mukhopadhyay, A.G, Toropova, K, Daly, L, Wells, J, Vuolo, L, Seda, M, Jenkins, D, Stephens, D.J, Roberts, A.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Embo J., 43, 2024
|
|
7V5L
 
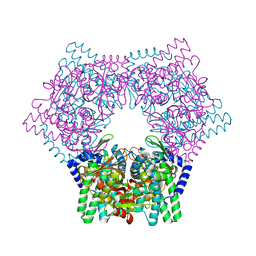 | | Crystal structure of human bleomycin hydrolase | | Descriptor: | Bleomycin hydrolase | | Authors: | Chang, C.Y, Zheng, Y.Z, Huang, S.J, Wang, Y.L, Toh, S.I, Lin, E.C. | | Deposit date: | 2021-08-17 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure-Function Relationship of Human Bleomycin Hydrolase: Mutation of a Cysteine Protease into a Serine Protease.
Chembiochem, 23, 2022
|
|
6CWR
 
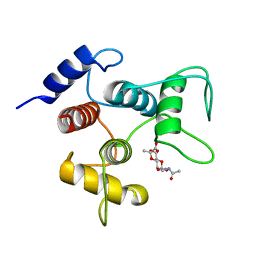 | | Crystal structure of SpaA-SLH/G46A/G109A in complex with 4,6-Pyr-beta-D-ManNAcOMe | | Descriptor: | Surface (S-) layer glycoprotein, methyl 2-(acetylamino)-4,6-O-[(1S)-1-carboxyethylidene]-2-deoxy-beta-D-mannopyranoside | | Authors: | Blackler, R.J, Gagnon, S.M.L, Haji-Ghassemi, O, Evans, S.V. | | Deposit date: | 2018-03-30 | | Release date: | 2018-08-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Structural basis of cell wall anchoring by SLH domains in Paenibacillus alvei.
Nat Commun, 9, 2018
|
|
5LHI
 
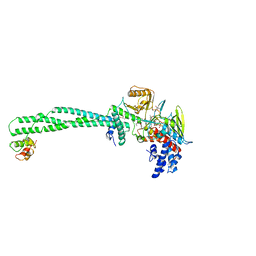 | | Structure of the KDM1A/CoREST complex with the inhibitor N-[3-(ethoxymethyl)-2-[[4-[[(3R)-pyrrolidin-3-yl]methoxy]phenoxy]methyl]phenyl]-4-methylthieno[3,2-b]pyrrole-5-carboxamide | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, LYSINE-SPECIFIC HISTONE DEMETHYLASE 1, ... | | Authors: | Cecatiello, V, Pasqualato, S. | | Deposit date: | 2016-07-12 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Thieno[3,2-b]pyrrole-5-carboxamides as New Reversible Inhibitors of Histone Lysine Demethylase KDM1A/LSD1. Part 2: Structure-Based Drug Design and Structure-Activity Relationship.
J. Med. Chem., 60, 2017
|
|
6GB2
 
 | | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. This file contains the 39S ribosomal subunit. | | Descriptor: | 'Mitochondrial ribosomal protein L30, 'Mitochondrial ribosomal protein L55, 'Mitochondrial ribosomal protein L59, ... | | Authors: | Kummer, E, Leibundgut, M, Boehringer, D, Ban, N. | | Deposit date: | 2018-04-13 | | Release date: | 2018-08-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM.
Nature, 560, 2018
|
|
7OHA
 
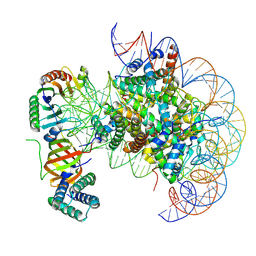 | | nucleosome with TBP and TFIIA bound at SHL +2 | | Descriptor: | DNA (122-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7SWY
 
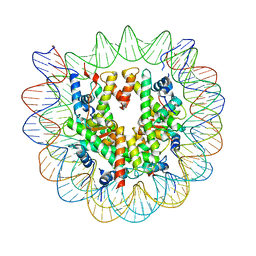 | | 2.6 A structure of a 40-601[TA-rich+1]-40 nucleosome | | Descriptor: | DNA Guide Strand, DNA Tracking Strand, Histone H2A, ... | | Authors: | Nodelman, I.M, Bowman, G.D, Armache, J.-P. | | Deposit date: | 2021-11-21 | | Release date: | 2022-03-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Nucleosome recognition and DNA distortion by the Chd1 remodeler in a nucleotide-free state.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OH9
 
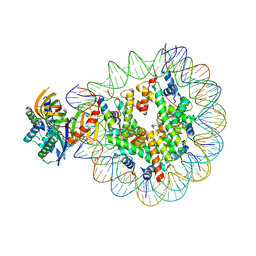 | | Nucleosome with TBP and TFIIA bound at SHL -6 | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-09 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7OHC
 
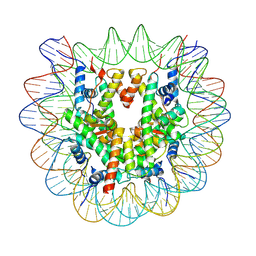 | |
7OHB
 
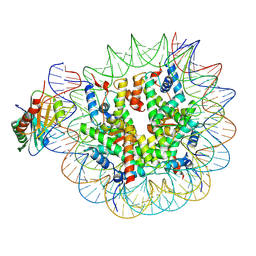 | | TBP-nucleosome complex | | Descriptor: | DNA (145-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Wang, H, Cramer, P. | | Deposit date: | 2021-05-10 | | Release date: | 2021-07-28 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures and implications of TBP-nucleosome complexes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3G5T
 
 | | Crystal structure of trans-aconitate 3-methyltransferase from yeast | | Descriptor: | (2E)-2-(2-methoxy-2-oxoethyl)but-2-enedioic acid, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Burgie, E.S, Bingman, C.A, Wesenberg, G.E, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-02-05 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | Crystal structure of trans-aconitate 3-methyltransferase from yeast
To be Published
|
|
6IV8
 
 | | the selenomethionine(SeMet)-derived Cas13d binary complex | | Descriptor: | MAGNESIUM ION, RNA (51-MER), RNA (53-MER), ... | | Authors: | Zhang, B, Ye, Y.M, Ye, W.W, OuYang, S.Y. | | Deposit date: | 2018-12-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Two HEPN domains dictate CRISPR RNA maturation and target cleavage in Cas13d.
Nat Commun, 10, 2019
|
|
1UGY
 
 | | Crystal structure of jacalin- mellibiose (Gal-alpha(1-6)-Glc) complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, alpha-D-galactopyranose-(1-6)-alpha-D-glucopyranose, ... | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-23 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
1W98
 
 | | The structural basis of CDK2 activation by cyclin E | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, G1/S-SPECIFIC CYCLIN E1 | | Authors: | Lowe, E.D, Honda, R, Dubinina, E, Skamnaki, V, Cook, A, Johnson, L.N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-02-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of Cyclin E1/Cdk2: Implications for Cdk2 Activation and Cdk2-Independent Roles
Embo J., 24, 2005
|
|
1UGX
 
 | | Crystal structure of jacalin- Me-alpha-T-antigen (Gal-beta(1-3)-GalNAc-alpha-o-Me) complex | | Descriptor: | Agglutinin alpha chain, Agglutinin beta-3 chain, beta-D-galactopyranose-(1-3)-methyl 2-acetamido-2-deoxy-alpha-D-galactopyranoside | | Authors: | Jeyaprakash, A.A, Katiyar, S, Swaminathan, C.P, Sekar, K, Surolia, A, Vijayan, M. | | Deposit date: | 2003-06-22 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis of the Carbohydrate Specificities of Jacalin: An X-ray and Modeling Study
J.MOL.BIOL., 332, 2003
|
|
1W1D
 
 | | Crystal Structure of the PDK1 Pleckstrin Homology (PH) domain bound to Inositol (1,3,4,5)-tetrakisphosphate | | Descriptor: | 3-PHOSPHOINOSITIDE DEPENDENT PROTEIN KINASE-1, GLYCEROL, GOLD ION, ... | | Authors: | Komander, D, Deak, M, Alessi, D.R, Van Aalten, D.M.F. | | Deposit date: | 2004-06-21 | | Release date: | 2004-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Insights Into the Regulation of Pdk1 by Phosphoinositides and Inositol Phosphates
Embo J., 23, 2004
|
|
3IHR
 
 | | Crystal Structure of Uch37 | | Descriptor: | FORMIC ACID, SODIUM ION, Ubiquitin carboxyl-terminal hydrolase isozyme L5 | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-11 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural characterization of human Uch37.
Proteins, 80, 2012
|
|
6ROI
 
 | | Cryo-EM structure of the partially activated Drs2p-Cdc50p | | Descriptor: | (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Timcenko, M, Lyons, J.A, Januliene, D, Ulstrup, J.J, Dieudonne, T, Montigny, C, Ash, M.R, Karlsen, J.L, Boesen, T, Kuhlbrandt, W, Lenoir, G, Moeller, A, Nissen, P. | | Deposit date: | 2019-05-13 | | Release date: | 2019-07-03 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure and autoregulation of a P4-ATPase lipid flippase.
Nature, 571, 2019
|
|
