6N17
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ577 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(3-carboxypropanoyl)amino]benzene-1,2-dicarboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-08 | | Release date: | 2019-07-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.639 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
6N0R
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ572 | | Descriptor: | 1,2-ETHANEDIOL, 4-(methylamino)benzene-1,2-dicarboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-11-07 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.544 Å) | | Cite: | Crystal structure of Tdp1 catalytic domain
To Be Published
|
|
1P9L
 
 | | Structure of M. tuberculosis dihydrodipicolinate reductase in complex with NADH and 2,6 PDC | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PYRIDINE-2,6-DICARBOXYLIC ACID, TETRAETHYLENE GLYCOL, ... | | Authors: | Cirilli, M, Zheng, R, Scapin, G, Blanchard, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-05-12 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The three-dimensional structures of the Mycobacterium tuberculosis dihydrodipicolinate reductase-NADH-2,6-PDC and -NADPH-2,6-PDC complexes. Structural and mutagenic analysis of relaxed nucleotide specificity.
Biochemistry, 42, 2003
|
|
4BQN
 
 | | Structural insights into WcbI, a novel polysaccharide biosynthesis enzyme. Native protein. | | Descriptor: | CAPSULAR POLYSACCHARIDE BIOSYNTHESIS PROTEIN, CHLORIDE ION, COENZYME A, ... | | Authors: | Vivoli, M, Ayres, E, Isupov, M.N, Harmer, N.J. | | Deposit date: | 2013-05-31 | | Release date: | 2013-11-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Structural Insights Into Wcbi, a Novel Polysaccharide-Biosynthesis Enzyme.
Iucrj, 1, 2014
|
|
4BQO
 
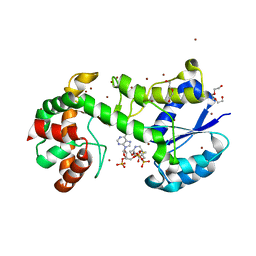 | | Structural insights into WcbI, a novel polysaccharide biosynthesis enzyme. Native protein without disulfide bond between COA and Cys14. | | Descriptor: | BROMIDE ION, COENZYME A, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vivoli, M, Ayres, E, Isupov, M.N, Harmer, N.J. | | Deposit date: | 2013-05-31 | | Release date: | 2013-11-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural Insights Into Wcbi, a Novel Polysaccharide-Biosynthesis Enzyme.
Iucrj, 1, 2014
|
|
4TX3
 
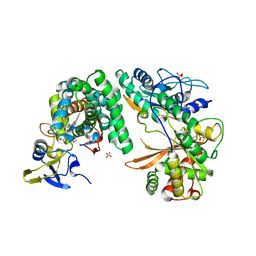 | | Complex of the X-domain and OxyB from Teicoplanin Biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, OxyB protein, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Peschke, M, Haslinger, K, Cryle, M.J. | | Deposit date: | 2014-07-02 | | Release date: | 2015-02-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-domain of peptide synthetases recruits oxygenases crucial for glycopeptide biosynthesis.
Nature, 521, 2015
|
|
5SD3
 
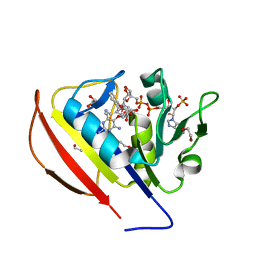 | |
5SD6
 
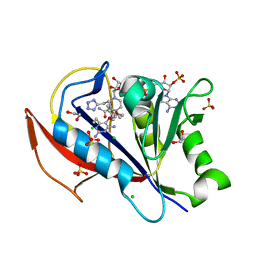 | |
2JRE
 
 | | C60-1, a PDZ domain designed using statistical coupling analysis | | Descriptor: | C60-1 PDZ domain peptide | | Authors: | Larson, C, Stiffler, M, Li, P, Rosen, M, MacBeath, G, Ranganathan, R. | | Deposit date: | 2007-06-25 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | C60-1, a PDZ domain designed using statistical coupling analysis
To be Published
|
|
2A5P
 
 | | Monomeric parallel-stranded DNA tetraplex with snap-back 3+1 3' G-tetrad, single-residue chain reversal loops, GAG triad in the context of GAAG diagonal loop, NMR, 8 struct. | | Descriptor: | 5'-D(*TP*GP*AP*GP*GP*GP*TP*GP*GP*IP*GP*AP*GP*GP*GP*TP*GP*GP*GP*GP*AP*AP*GP*G)-3' | | Authors: | Phan, A.T, Kuryavyi, V.V, Gaw, H.Y, Patel, D.J. | | Deposit date: | 2005-06-30 | | Release date: | 2005-07-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Small-molecule interaction with a five-guanine-tract G-quadruplex structure from the human MYC promoter.
Nat.Chem.Biol., 1, 2005
|
|
1JQA
 
 | | Bacillus stearothermophilus glycerol dehydrogenase complex with glycerol | | Descriptor: | GLYCEROL, Glycerol Dehydrogenase, ZINC ION | | Authors: | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | Deposit date: | 2001-08-04 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
6D3U
 
 | | Complex structure of Ulvan lyase from Nonlaben Ulvanivorans- NLR48 | | Descriptor: | 1,2-ETHANEDIOL, 4-deoxy-alpha-L-threo-hex-4-enopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-3-O-sulfo-alpha-L-rhamnopyranose, CALCIUM ION, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-06 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structural and functional characterization of PL28 family ulvan lyase NLR48 fromNonlabens ulvanivorans.
J. Biol. Chem., 293, 2018
|
|
6JIB
 
 | | Human MTHFD2 in complex with DS44960156 | | Descriptor: | 4-(5-oxo-1,5-dihydro-2H-[1]benzopyrano[3,4-c]pyridine-3(4H)-carbonyl)benzoic acid, Bifunctional methylenetetrahydrofolate dehydrogenase/cyclohydrolase, mitochondrial, ... | | Authors: | Suzuki, M, Matsui, Y, Kawai, J. | | Deposit date: | 2019-02-20 | | Release date: | 2019-06-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-Based Design and Synthesis of an Isozyme-Selective MTHFD2 Inhibitor with a Tricyclic Coumarin Scaffold.
Acs Med.Chem.Lett., 10, 2019
|
|
6JS8
 
 | | Structure of the CYP102A1 Haem Domain with N-Dehydroabietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aS,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,9,10,10a-hexahydrophenanthren-1-yl]carbonylamino]-3-(1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Stanfield, J.K, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-04-07 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
2YVJ
 
 | | Crystal structure of the ferredoxin-ferredoxin reductase (BPHA3-BPHA4)complex | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Biphenyl dioxygenase ferredoxin subunit, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Senda, T, Senda, M. | | Deposit date: | 2007-04-12 | | Release date: | 2007-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Mechanism of the Redox-dependent Interaction between NADH-dependent Ferredoxin Reductase and Rieske-type [2Fe-2S] Ferredoxin
J.Mol.Biol., 373, 2007
|
|
5CIH
 
 | |
5GM9
 
 | | Crystal structure of a glycoside hydrolase in complex with cellobiose | | Descriptor: | Glycoside hydrolase family 45 protein, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Gao, J, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-13 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Characterization and crystal structure of a thermostable glycoside hydrolase family 45 1,4-beta-endoglucanase from Thielavia terrestris
Enzyme Microb. Technol., 99, 2017
|
|
5IED
 
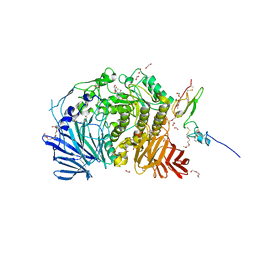 | | Murine endoplasmic reticulum alpha-glucosidase II with castanospermine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3CL9
 
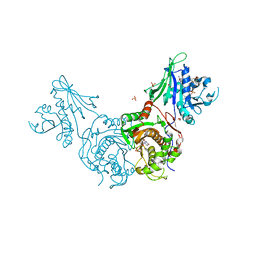 | | Structure of bifunctional TcDHFR-TS in complex with MTX | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, Bifunctional dihydrofolate reductase-thymidylate synthase (DHFR-TS), ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2008-03-18 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
3SZO
 
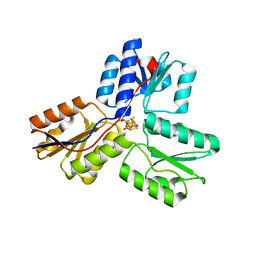 | | IspH:HMBPP complex after 3 minutes X-ray pre-exposure | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, 4-hydroxy-3-methylbut-2-enyl diphosphate reductase, IRON/SULFUR CLUSTER | | Authors: | Span, I, Graewert, T, Bacher, A, Eisenreich, W, Groll, M. | | Deposit date: | 2011-07-19 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Mutant IspH Proteins Reveal a Rotation of the Substrate's Hydroxymethyl Group during Catalysis.
J.Mol.Biol., 416, 2012
|
|
5IK8
 
 | | Laminin A2LG45 I-form, G6/7 bound. | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-hydroxy-4-methyl-2H-chromen-2-one, ... | | Authors: | Briggs, D.C, Hohenester, E, Campbell, K.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of laminin binding to the LARGE glycans on dystroglycan.
Nat.Chem.Biol., 12, 2016
|
|
6A0R
 
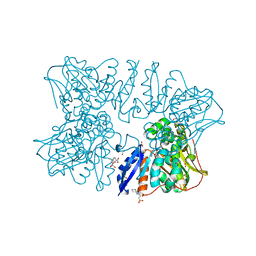 | | Homoserine dehydrogenase from Thermus thermophilus HB8 unliganded form | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Akai, S, Ikushiro, H, Sawai, T, Yano, T, Kamiya, N, Miyahara, I. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | The crystal structure of homoserine dehydrogenase complexed with l-homoserine and NADPH in a closed form
J. Biochem., 165, 2019
|
|
4ME3
 
 | | 1.8 Angstrom Crystal Structure of the N-terminal Domain of an Archaeal MCM | | Descriptor: | DNA replication licensing factor MCM related protein, ZINC ION | | Authors: | Fu, Y, Slaymaker, I.M, Wang, G, Chen, X.S. | | Deposit date: | 2013-08-24 | | Release date: | 2014-01-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.794 Å) | | Cite: | The 1.8- angstrom Crystal Structure of the N-Terminal Domain of an Archaeal MCM as a Right-Handed Filament.
J.Mol.Biol., 426, 2014
|
|
6A0S
 
 | | Homoserine dehydrogenase from Thermus thermophilus HB8 complexed with HSE and NADPH | | Descriptor: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, FORMIC ACID, GLYCEROL, ... | | Authors: | Akai, S, Ikushiro, H, Sawai, T, Yano, T, Kamiya, N, Miyahara, I. | | Deposit date: | 2018-06-06 | | Release date: | 2018-11-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of homoserine dehydrogenase complexed with l-homoserine and NADPH in a closed form
J. Biochem., 165, 2019
|
|
5GUG
 
 | | Crystal structure of inositol 1,4,5-trisphosphate receptor large cytosolic domain with inositol 1,4,5-trisphosphate | | Descriptor: | D-MYO-INOSITOL-1,4,5-TRIPHOSPHATE, Inositol 1,4,5-trisphosphate receptor type 1 | | Authors: | Hamada, K, Miyatake, H, Terauchi, A, Mikoshiba, K. | | Deposit date: | 2016-08-29 | | Release date: | 2017-04-26 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (7.399 Å) | | Cite: | IP3-mediated gating mechanism of the IP3 receptor revealed by mutagenesis and X-ray crystallography
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
