5ZTC
 
 | |
8VWT
 
 | |
8VWV
 
 | |
6NQA
 
 | | Active state Dot1L bound to the H2B-Ubiquitinated nucleosome, 1-to-1 complex | | Descriptor: | 601 DNA Strand 1, 601 DNA Strand 2, Histone H2A type 1, ... | | Authors: | Worden, E.J, Hoffmann, N.A, Wolberger, C. | | Deposit date: | 2019-01-19 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of Cross-talk between H2B Ubiquitination and H3 Methylation by Dot1L.
Cell, 176, 2019
|
|
5F4N
 
 | | Multi-parameter lead optimization to give an oral CHK1 inhibitor clinical candidate: (R)-5-((4-((morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Serine/threonine-protein kinase Chk1, ... | | Authors: | Collins, I, Garrett, M.D, van Montfort, R, Osborne, J.D, Matthews, T.P, McHardy, T, Proisy, N, Cheung, K.J, Lainchbury, M, Brown, N, Walton, M.I, Eve, P.D, Boxall, K.J, Hayes, A, Henley, A.T, Valenti, M.R, De Haven Brandon, A.K, Box, G, Westwood, I.M, Jamin, Y, Robinson, S.P, Leonard, P, Reader, J.C, Aherne, G.W, Raynaud, F.I, Eccles, S.A. | | Deposit date: | 2015-12-03 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Multiparameter Lead Optimization to Give an Oral Checkpoint Kinase 1 (CHK1) Inhibitor Clinical Candidate: (R)-5-((4-((Morpholin-2-ylmethyl)amino)-5-(trifluoromethyl)pyridin-2-yl)amino)pyrazine-2-carbonitrile (CCT245737).
J.Med.Chem., 59, 2016
|
|
4MH8
 
 | |
8IUD
 
 | |
8IMW
 
 | |
4Y13
 
 | | SdiA in complex with octanoyl-rac-glycerol | | Descriptor: | (2S)-2,3-dihydroxypropyl octanoate, GLYCEROL, SULFATE ION, ... | | Authors: | Nguyen, N.X, Nguyen, Y, Sperandio, V, Jiang, Y. | | Deposit date: | 2015-02-06 | | Release date: | 2015-04-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Structural and Mechanistic Roles of Novel Chemical Ligands on the SdiA Quorum-Sensing Transcription Regulator.
Mbio, 6, 2015
|
|
8G6Q
 
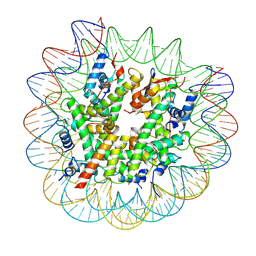 | | H2AK119ub-modified nucleosome ubiquitin position 1 | | Descriptor: | 601 DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Hicks, C.W, Wolberger, C, Keogh, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Ubiquitinated histone H2B as gatekeeper of the nucleosome acidic patch.
Nucleic Acids Res., 52, 2024
|
|
8G6S
 
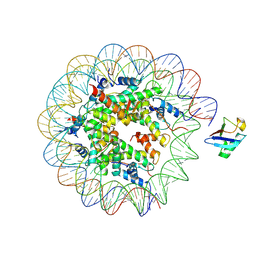 | | H2AK119ub-modified nucleosome ubiquitin position 2 | | Descriptor: | 601 DNA (147-MER), Histone H2A, Histone H2B 1.1, ... | | Authors: | Hicks, C.W, Wolberger, C, Keogh, M. | | Deposit date: | 2023-02-15 | | Release date: | 2024-02-21 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Ubiquitinated histone H2B as gatekeeper of the nucleosome acidic patch.
Nucleic Acids Res., 52, 2024
|
|
1PVU
 
 | |
7UJ1
 
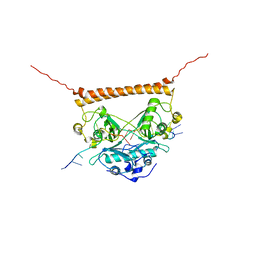 | | Crystal structure of PSF-RNA complex | | Descriptor: | RNA (30-MER), Splicing factor, proline- and glutamine-rich | | Authors: | Sachpatzidis, A, Wang, J, Konigsberg, W.H. | | Deposit date: | 2022-03-30 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insight into the Tumor Suppression Mechanism from the Structure of Human Polypyrimidine Splicing Factor (PSF/SFPQ) Complexed with a 30mer RNA from Murine Virus-like 30S Transcript-1.
Biochemistry, 61, 2022
|
|
4DRA
 
 | | Crystal structure of MHF complex | | Descriptor: | Centromere protein S, Centromere protein X | | Authors: | Tao, Y, Niu, L, Teng, M. | | Deposit date: | 2012-02-17 | | Release date: | 2012-05-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.414 Å) | | Cite: | The structure of the FANCM-MHF complex reveals physical features for functional assembly
Nat Commun, 3, 2012
|
|
1LJ9
 
 | | The crystal structure of the transcriptional regulator SlyA | | Descriptor: | transcriptional regulator SlyA | | Authors: | Wu, R.Y, Zhang, R.G, Gornicki, P, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-04-19 | | Release date: | 2003-01-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Enterococcus faecalis SlyA-like transcriptional factor
J.Biol.Chem., 278, 2003
|
|
3P56
 
 | |
1XP8
 
 | | Deinococcus radiodurans RecA in complex with ATP-gamma-S | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, RecA protein | | Authors: | Bell, C.E, Rajan, R. | | Deposit date: | 2004-10-08 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of RecA from Deinococcus radiodurans: insights into the structural basis of extreme radioresistance.
J.Mol.Biol., 344, 2004
|
|
2W42
 
 | | THE STRUCTURE OF A PIWI PROTEIN FROM ARCHAEOGLOBUS FULGIDUS COMPLEXED WITH A 16NT DNA DUPLEX. | | Descriptor: | 5'-D(*GP*TP*CP*GP*AP*AP*TP*TP)-3', 5'-D(*TP*TP*CP*GP*AP*CP*GP*CP)-3', MANGANESE (II) ION, ... | | Authors: | Parker, J.S, Roe, S.M, Barford, D. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Enhancement of the Seed-Target Recognition Step in RNA Silencing by a Piwi-Mid Domain Protein
Mol.Cell, 33, 2009
|
|
7K7T
 
 | | Crystal structure of human MORC4 ATPase-CW in complex with AMPPNP | | Descriptor: | Isoform 3 of MORC family CW-type zinc finger protein 4, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Klein, B.J, Tencer, A.H, Kutateladze, T.G. | | Deposit date: | 2020-09-24 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Molecular mechanism of the MORC4 ATPase activation.
Nat Commun, 11, 2020
|
|
7VII
 
 | |
7VIK
 
 | |
4WWX
 
 | | Crystal structure of the core RAG1/2 recombinase | | Descriptor: | V(D)J recombination-activating protein 1, V(D)J recombination-activating protein 2, ZINC ION | | Authors: | Kim, M.S, Lapkouski, M, Yang, W, Gellert, M. | | Deposit date: | 2014-11-12 | | Release date: | 2015-02-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.2001 Å) | | Cite: | Crystal structure of the V(D)J recombinase RAG1-RAG2.
Nature, 518, 2015
|
|
2HVS
 
 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 2'-deoxyribonucleotide at the nick | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
2HVR
 
 | | Structure of T4 RNA Ligase 2 with Nicked 5'-Adenylated nucleic acid duplex containing a 3'-deoxyribonucleotide at the nick | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-D(*AP*TP*TP*CP*CP*GP*AP*TP*AP*GP*TP*GP*GP*GP*GP*TP*CP*GP*CP*AP*AP*TP*TP*G)-3', 5'-D(*CP*AP*AP*TP*TP*GP*CP*GP*AP*C)-R(P*(OMC)P*C)-3', ... | | Authors: | Nandakumar, J, Lima, C.D. | | Deposit date: | 2006-07-30 | | Release date: | 2006-10-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | RNA Ligase Structures Reveal the Basis for RNA Specificity and Conformational Changes that Drive Ligation Forward.
Cell(Cambridge,Mass.), 127, 2006
|
|
8A1H
 
 | | Bacterial 6-4 photolyase from Vibrio cholerase | | Descriptor: | 1-deoxy-1-(6,7-dimethyl-2,4-dioxo-3,4-dihydropteridin-8(2H)-yl)-D-ribitol, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 6-4 photolyase (FeS-BCP, ... | | Authors: | Essen, L.-O, Emmerich, H.J. | | Deposit date: | 2022-06-01 | | Release date: | 2023-02-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural and Functional Analysis of a Prokaryotic (6-4) Photolyase from the Aquatic Pathogen Vibrio Cholerae † .
Photochem.Photobiol., 99, 2023
|
|
