8GMM
 
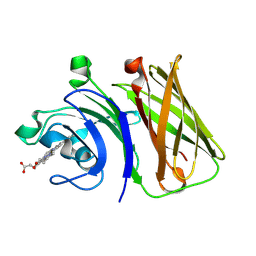 | | Stenotrophomonas maltophilia Holo HphA | | Descriptor: | HEME B/C, Hemophilin | | Authors: | Shin, H.E, Moraes, T.F. | | Deposit date: | 2023-03-26 | | Release date: | 2024-03-27 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Prevalence of Slam-dependent hemophilins in Gram-negative bacteria.
J.Bacteriol., 206, 2024
|
|
6N6N
 
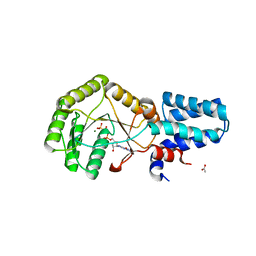 | | FtsY-NG high-resolution | | Descriptor: | ACETATE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ataide, S.F, Faoro, C. | | Deposit date: | 2018-11-26 | | Release date: | 2019-10-09 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.877 Å) | | Cite: | Structural insights into the G-loop dynamics of E. coli FtsY NG domain.
J.Struct.Biol., 208, 2019
|
|
5DIQ
 
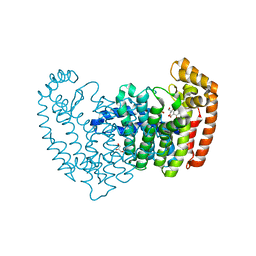 | | Crystal structure of human FPPS in complex with salicylic acid derivative 3a | | Descriptor: | 2-(naphthalen-1-ylmethoxy)benzoic acid, Farnesyl pyrophosphate synthase, GLYCEROL, ... | | Authors: | Rondeau, J.M, Bourgier, E, Lehmann, S. | | Deposit date: | 2015-09-01 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Novel Allosteric Non-Bisphosphonate Inhibitors of Farnesyl Pyrophosphate Synthase by Integrated Lead Finding.
Chemmedchem, 10, 2015
|
|
4RGZ
 
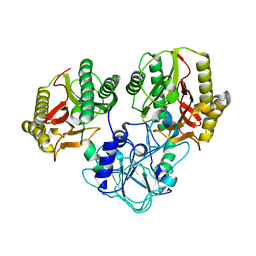 | | Crystal structure of recombinant prolidase from Thermococcus sibiricus at P21221 spacegroup | | Descriptor: | PHOSPHATE ION, Xaa-Pro aminopeptidase, ZINC ION | | Authors: | Timofeev, V.I, Korgenevsky, D.A, Gorbacheva, M.A, Boyko, K.M, Slutsky, E, Rakitina, T.V, Lipkin, A.V, Popov, V.O. | | Deposit date: | 2014-10-01 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure of recombinant prolidase from Thermococcus sibiricus in space group P21221.
Acta Crystallogr.,Sect.F, 71, 2015
|
|
5LQK
 
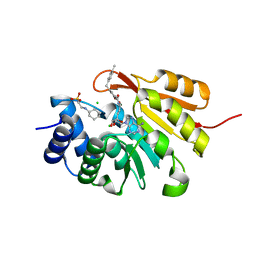 | | Crystal Structure of COMT in complex with N-[(E)-3-[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxyoxolan-2-yl]prop-2-enyl]-2,3-dihydroxy-5-[(4-methylphenyl)methyl]benzamide | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CHLORIDE ION, Catechol O-methyltransferase, ... | | Authors: | Ehler, A, Lerner, C, Rudolph, M.G. | | Deposit date: | 2016-08-17 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of COMT
To be published
|
|
2JAA
 
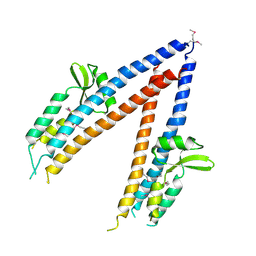 | | SeMet substituted Shigella Flexneri Ipad | | Descriptor: | INVASIN IPAD | | Authors: | Johnson, S, Roversi, P, Espina, M, Olive, A, Deane, J.E, Birket, S, Field, T, Picking, W.D, Blocker, A.J, Galyov, E.E, Picking, W.L, Lea, S.M. | | Deposit date: | 2006-11-24 | | Release date: | 2006-11-30 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Self-Chaperoning of the Type III Secretion System Needle Tip Proteins Ipad and Bipd.
J.Biol.Chem., 282, 2007
|
|
1FM4
 
 | | CRYSTAL STRUCTURE OF THE BIRCH POLLEN ALLERGEN BET V 1L | | Descriptor: | (3ALPHA,5BETA,12ALPHA)-3,12-DIHYDROXYCHOLAN-24-OIC ACID, MAJOR POLLEN ALLERGEN BET V 1-L | | Authors: | Markovic-Housley, Z, Degano, M, Lamba, D, von Roepenack-Lahaye, E, Clemens, S, Susani, M, Ferreira, F, Scheiner, O, Breiteneder, H. | | Deposit date: | 2000-08-16 | | Release date: | 2002-12-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal Structure of a Hypoallergenic Isoform of the Major Birch Pollen Allergen Bet
v 1 and its Likely Biological Function as a Plant Steroid Carrier
J.Mol.Biol., 325, 2003
|
|
1ABT
 
 | |
1AC7
 
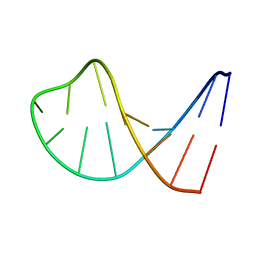 | | STRUCTURAL FEATURES OF THE DNA HAIRPIN D(ATCCTAGTTATAGGAT): THE FORMATION OF A G-A BASE PAIR IN THE LOOP, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*TP*AP*GP*TP*TP*AP*TP*AP*GP*GP*AP*T)-3') | | Authors: | Van Dongen, M.J.P, Mooren, M.M.W, Willems, E.F.A, Van Der Marel, G.A, Van Boom, J.H, Wijmenga, S.S, Hilbers, C.W. | | Deposit date: | 1997-02-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural features of the DNA hairpin d(ATCCTA-GTTA-TAGGAT): formation of a G-A base pair in the loop.
Nucleic Acids Res., 25, 1997
|
|
8TBQ
 
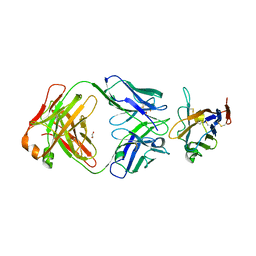 | |
5ECW
 
 | |
4L6D
 
 | | Crystal structure of 5-carboxyvanillate decarboxylase from Sphingomonas paucimobilis complexed with vanillic acid | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-HYDROXY-3-METHOXYBENZOATE, 5-carboxyvanillate decarboxylase, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Vladimirova, A, Raushel, F.M, Almo, S.C. | | Deposit date: | 2013-06-12 | | Release date: | 2013-11-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Crystal structure of 5-carboxyvanillate decarboxylase from Sphingomonas paucimobilis complexed with vanillic acid
To be Published
|
|
6NC1
 
 | | FtsY-NG high-resolution | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Ataide, S.F, Faoro, C. | | Deposit date: | 2018-12-10 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Structural insights into the G-loop dynamics of E. coli FtsY NG domain.
J.Struct.Biol., 208, 2019
|
|
5LVL
 
 | | Human PDK1 Kinase Domain in Complex with Compound PS653 Bound to the ATP-Binding Site | | Descriptor: | 2,6-DIHYDROANTHRA/1,9-CD/PYRAZOL-6-ONE, 3-phosphoinositide-dependent protein kinase 1, DIMETHYL SULFOXIDE, ... | | Authors: | Schulze, J.O, Saladino, G, Busschots, K, Neimanis, S, Suess, E, Odadzic, D, Zeuzem, S, Hindie, V, Herbrand, A.K, Lisa, M.N, Alzari, P.M, Gervasio, F.L, Biondi, R.M. | | Deposit date: | 2016-09-14 | | Release date: | 2016-10-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Bidirectional Allosteric Communication between the ATP-Binding Site and the Regulatory PIF Pocket in PDK1 Protein Kinase.
Cell Chem Biol, 23, 2016
|
|
7UIT
 
 | |
2VZC
 
 | | Crystal structure of the C-terminal calponin homology domain of alpha parvin | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Lorenz, S, Vakonakis, I, Lowe, E.D, Campbell, I.D, Noble, M.E.M, Hoellerer, M.K. | | Deposit date: | 2008-07-31 | | Release date: | 2008-10-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural Analysis of the Interactions between Paxillin Ld Motifs and Alpha-Parvin
Structure, 16, 2008
|
|
4IVZ
 
 | |
4Y1W
 
 | |
5EI0
 
 | | Structure of RCL-cleaved vaspin (serpinA12) | | Descriptor: | Serpin A12 | | Authors: | Pippel, J, Kuettner, B.E, Ulbricht, D, Daberger, J, Schultz, S, Heiker, J.T, Strater, N. | | Deposit date: | 2015-10-29 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of cleaved vaspin (serpinA12).
Biol.Chem., 397, 2016
|
|
2CNI
 
 | | Structural Insights into the Design of Nonpeptidic Isothiazolidinone- Containing Inhibitors of Protein Tyrosine Phosphatase 1B | | Descriptor: | MAGNESIUM ION, METHYL 2-{[5-({3-CHLORO-4-[(5S)-1,1-DIOXIDO-3-OXOISOTHIAZOLIDIN-5-YL]-N-(PHENYLSULFONYL)-L-PHENYLALANYL}AMINO)PENTYL]OXY}-6-HYDROXYBENZOATE, TYROSINE-PROTEIN PHOSPHATASE NON-RECEPTOR TYPE 1 | | Authors: | Ala, P.J, Gonneville, L, Hillman, M, Becker-Pasha, M, Yue, E.W, Douty, B, Wayland, B, Polam, P, Crawley, M.L, McLaughlin, E, Sparks, R.B, Glass, B, Takvorian, A, Combs, A.P, Burn, T.C, Hollis, G.F, Wynn, R. | | Deposit date: | 2006-05-21 | | Release date: | 2006-09-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights Into the Design of Nonpeptidic Isothiazolidinone-Containing Inhibitors of Protein- Tyrosine Phosphatase 1B.
J.Biol.Chem., 281, 2006
|
|
1OEG
 
 | | PEPTIDE OF HUMAN APOE RESIDUES 267-289, NMR, 5 STRUCTURES AT PH 6.0, 37 DEGREES CELSIUS AND PEPTIDE:SDS MOLE RATIO OF 1:90 | | Descriptor: | APOLIPOPROTEIN E | | Authors: | Wang, G, Pierens, G.K, Treleaven, W.D, Sparrow, J.T, Cushley, R.J. | | Deposit date: | 1996-03-16 | | Release date: | 1996-12-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformations of human apolipoprotein E(263-286) and E(267-289) in aqueous solutions of sodium dodecyl sulfate by CD and 1H NMR.
Biochemistry, 35, 1996
|
|
1HXE
 
 | | SERINE PROTEASE | | Descriptor: | HIRUDIN VARIANT-1, RUBIDIUM ION, THROMBIN | | Authors: | Tulinsky, A, Zhang, E. | | Deposit date: | 1995-12-07 | | Release date: | 1996-11-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The molecular environment of the Na+ binding site of thrombin.
Biophys.Chem., 63, 1997
|
|
4IPY
 
 | | HIV capsid C-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein p24 | | Authors: | Lampel, A, Yaniv, O, Berger, O, Bachrach, E, Gazit, E, Frolow, F. | | Deposit date: | 2013-01-10 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A triclinic crystal structure of the carboxy-terminal domain of HIV-1 capsid protein with four molecules in the asymmetric unit reveals a novel packing interface.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
1HL6
 
 | | A novel mode of RBD-protein recognition in the Y14-mago complex | | Descriptor: | CG8781 PROTEIN, MAGO NASHI PROTEIN | | Authors: | Fribourg, S, Gatfield, D, Yao, W, Izaurralde, E, Conti, E. | | Deposit date: | 2003-03-13 | | Release date: | 2003-05-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A Novel Mode of Rbd-Protein Recognition in the Y14-Mago Complex
Nat.Struct.Biol., 10, 2003
|
|
6BQG
 
 | | Crystal structure of 5-HT2C in complex with ergotamine | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 5-hydroxytryptamine receptor 2C,Soluble cytochrome b562, Ergotamine | | Authors: | Peng, Y, McCorvy, J.D, Harpsoe, K, Lansu, K, Yuan, S, Popov, P, Qu, L, Pu, M, Che, T, Nikolajse, L.F, Huang, X.P, Wu, Y, Shen, L, Bjorn-Yoshimoto, W.E, Ding, K, Wacker, D, Han, G.W, Cheng, J, Katritch, V, Jensen, A.A, Hanson, M.A, Zhao, S, Gloriam, D.E, Roth, B.L, Stevens, R.C, Liu, Z. | | Deposit date: | 2017-11-27 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 5-HT2C Receptor Structures Reveal the Structural Basis of GPCR Polypharmacology.
Cell, 172, 2018
|
|
