3CTN
 
 | | STRUCTURE OF CALCIUM-SATURATED CARDIAC TROPONIN C, NMR, 30 STRUCTURES | | Descriptor: | CALCIUM ION, TROPONIN C | | Authors: | Sia, S.K, Li, M.X, Spyracopoulos, L, Gagne, S.M, Liu, W, Putkey, J.A, Sykes, B.D. | | Deposit date: | 1997-05-08 | | Release date: | 1998-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of cardiac muscle troponin C unexpectedly reveals a closed regulatory domain.
J.Biol.Chem., 272, 1997
|
|
4KLE
 
 | | DNA polymerase beta matched reactant complex with Mg2+, 10 s | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Beard, W.A, Shock, D.D, Wilson, S.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Observing a DNA polymerase choose right from wrong.
Cell(Cambridge,Mass.), 154, 2013
|
|
4KLS
 
 | | DNA polymerase beta mismatched reactant complex with Mn2+, 10 min | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*A)-3', ... | | Authors: | Freudenthal, B.D, Beard, W.A, Shock, D.D, Wilson, S.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.978 Å) | | Cite: | Observing a DNA polymerase choose right from wrong.
Cell(Cambridge,Mass.), 154, 2013
|
|
4KLO
 
 | | DNA polymerase beta matched nick complex with Mg2+ and PPi, 30 min | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*GP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*CP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Freudenthal, B.D, Beard, W.A, Shock, D.D, Wilson, S.H. | | Deposit date: | 2013-05-07 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.845 Å) | | Cite: | Observing a DNA polymerase choose right from wrong.
Cell(Cambridge,Mass.), 154, 2013
|
|
3DDK
 
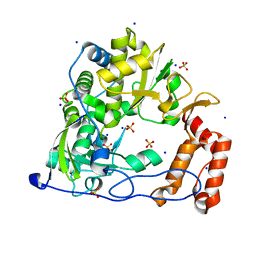 | | Coxsackievirus B3 3Dpol RNA Dependent RNA Polymerase | | Descriptor: | RNA polymerase B3 3Dpol, SODIUM ION, SULFATE ION | | Authors: | Campagnola, G, Weygandt, M.H, Scoggin, K.E, Peersen, O.B. | | Deposit date: | 2008-06-05 | | Release date: | 2008-09-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure of Coxsackievirus B3 3Dpol Highlights Functional Importance of Residue 5 in Picornaviral Polymerases
J.Virol., 82, 2008
|
|
3DDZ
 
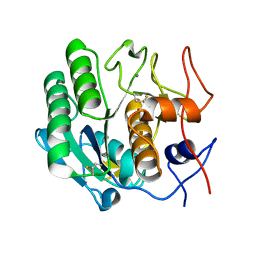 | |
7LPH
 
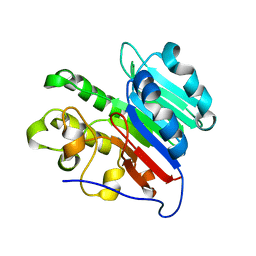 | | APE1 Mn-bound product complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-R(P*N)-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
7LPJ
 
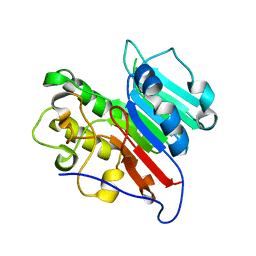 | | APE1 Mn-bound phosphorothioate substrate complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-R(P*(YA4))-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA-(apurinic or apyrimidinic site) lyase, ... | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
3D92
 
 | | Human carbonic anhydrase II bound with substrate carbon dioxide | | Descriptor: | CARBON DIOXIDE, GLYCEROL, ZINC ION, ... | | Authors: | Domsic, J.F, Avvaru, B.S, McKenna, R. | | Deposit date: | 2008-05-26 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Entrapment of carbon dioxide in the active site of carbonic anhydrase II
J.Biol.Chem., 283, 2008
|
|
7LPI
 
 | | APE1 phosphorothioate substrate complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-R(P*(YA4))-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
3DE1
 
 | |
7LPG
 
 | | APE1 product complex with abasic ribonucleotide DNA | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*GP*AP*TP*CP*CP*GP*TP*CP*GP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3'), DNA (5'-R(P*N)-D(P*CP*GP*AP*CP*GP*GP*AP*TP*CP*C)-3'), ... | | Authors: | Freudenthal, B.D, Hoitsma, N.M. | | Deposit date: | 2021-02-11 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Altered APE1 activity on abasic ribonucleotides is mediated by changes in the nucleoside sugar pucker.
Comput Struct Biotechnol J, 19, 2021
|
|
3DDU
 
 | | Prolyl Oligopeptidase with GSK552 | | Descriptor: | (6S)-1-chloro-3-[(4-fluorobenzyl)oxy]-6-(pyrrolidin-1-ylcarbonyl)pyrrolo[1,2-a]pyrazin-4(6H)-one, ACETATE ION, GLYCEROL, ... | | Authors: | Madauss, K.P, Reid, R.A, Haffner, C.D, Miller, A.B. | | Deposit date: | 2008-06-06 | | Release date: | 2008-08-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Pyrrolidinyl pyridone and pyrazinone analogues as potent inhibitors of prolyl oligopeptidase (POP)
Bioorg.Med.Chem.Lett., 18, 2008
|
|
5AYX
 
 | | Crystal structure of Human Quinolinate Phosphoribosyltransferase | | Descriptor: | Nicotinate-nucleotide pyrophosphorylase [carboxylating] | | Authors: | Kang, G.B, Kim, M.-K, Im, Y.J, Lee, J.H, Youn, H.-S, An, J.Y, Lee, J.-G, Fukuoka, S.-I, Eom, S.H. | | Deposit date: | 2015-09-14 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Quaternary Catalytic Mechanism of Hexameric Human Quinolinate Phosphoribosyltransferase, a Key Enzyme in de novo NAD Biosynthesis
Sci Rep, 6, 2016
|
|
2OVH
 
 | | Progesterone Receptor with Bound Asoprisnil and a Peptide from the Co-Repressor SMRT | | Descriptor: | 4-[(11BETA,17BETA)-17-METHOXY-17-(METHOXYMETHYL)-3-OXOESTRA-4,9-DIEN-11-YL]BENZALDEHYDE OXIME, Progesterone receptor, SMRT peptide | | Authors: | Madauss, K.P, Deng, S.-J, Short, S.A, Stewart, E.L, Williams, S.P. | | Deposit date: | 2007-02-13 | | Release date: | 2007-03-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A structural and in vitro characterization of asoprisnil: a selective progesterone receptor modulator.
Mol.Endocrinol., 21, 2007
|
|
3D9Q
 
 | |
4MGT
 
 | | ALKBH2 R110A cross-linked to undamaged dsDNA | | Descriptor: | Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, DNA1, DNA2, ... | | Authors: | Chen, B, Gan, J, Yang, C.G. | | Deposit date: | 2013-08-28 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The complex structures of ALKBH2 mutants cross-linked to
dsDNA reveal the conformational swing of β-hairpin
Sci China Chem, 57, 2014
|
|
3DE0
 
 | |
1US8
 
 | | The Rad50 signature motif: essential to ATP binding and biological function | | Descriptor: | DNA DOUBLE-STRAND BREAK REPAIR RAD50 ATPASE | | Authors: | Moncalian, G, Lengsfeld, B, Bhaskara, V, Hopfner, K.P, Karcher, A, Alden, E, Tainer, J.A, Paull, T.T. | | Deposit date: | 2003-11-20 | | Release date: | 2003-11-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Rad50 Signature Motif: Essential to ATP Binding and Biological Function
J.Mol.Biol., 335, 2004
|
|
4LOF
 
 | | Human p53 Core Domain Mutant V157F/N235K/N239Y | | Descriptor: | Cellular tumor antigen p53, ZINC ION | | Authors: | Wallentine, B.D, Wang, Y, Luecke, H. | | Deposit date: | 2013-07-12 | | Release date: | 2013-07-31 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of oncogenic, suppressor and rescued p53 core-domain variants: mechanisms of mutant p53 rescue.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3DE2
 
 | |
2OVV
 
 | | Crystal structure of the catalytic domain of rat phosphodiesterase 10A | | Descriptor: | 6,7-DIMETHOXY-4-[(3R)-3-(2-NAPHTHYLOXY)PYRROLIDIN-1-YL]QUINAZOLINE, MAGNESIUM ION, Phosphodiesterase-10A, ... | | Authors: | Pandit, J, Marr, E.S. | | Deposit date: | 2007-02-15 | | Release date: | 2007-03-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a series of 6,7-dimethoxy-4-pyrrolidylquinazoline PDE10A inhibitors
J.Med.Chem., 50, 2007
|
|
4LTT
 
 | | Crystal structure of native apo toxin from Helicobacter pylori | | Descriptor: | Uncharacterized protein, toxin | | Authors: | Lee, B.J, Im, H, Pathak, C.C, Yoon, H.J. | | Deposit date: | 2013-07-23 | | Release date: | 2014-02-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Crystal structure of apo and copper bound HP0894 toxin from Helicobacter pylori 26695 and insight into mRNase activity
Biochim.Biophys.Acta, 1834, 2013
|
|
4LUA
 
 | |
3D93
 
 | |
