3GW3
 
 | | human UROD mutant K297N | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Hill, C.P, Phillips, J.D, Whitby, F.G, Warby, C, Kushner, J.P. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and kinetic characterization of mutant human uroporphyrinogen decarboxylases.
Cell Mol Biol (Noisy-le-grand), 55, 2009
|
|
3GYZ
 
 | | Crystal structure of IpgC from Shigella flexneri | | Descriptor: | Chaperone protein ipgC, GLYCEROL, SODIUM ION, ... | | Authors: | Lunelli, M, Lokareddy, R.K, Zychlinsky, A, Kolbe, M. | | Deposit date: | 2009-04-06 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | IpaB-IpgC interaction defines binding motif for type III secretion translocator
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3GZ8
 
 | |
3GWK
 
 | | Structure of the homodimeric WXG-100 family protein from Streptococcus agalactiae | | Descriptor: | Putative uncharacterized protein SAG1039, SULFATE ION | | Authors: | Poulsen, C, Gries, F, Wilmanns, M, Song, Y.H. | | Deposit date: | 2009-04-01 | | Release date: | 2010-09-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | WXG100 protein superfamily consists of three subfamilies and exhibits an alpha-helical C-terminal conserved residue pattern.
Plos One, 9, 2014
|
|
3GZR
 
 | |
3GWZ
 
 | | Structure of the Mitomycin 7-O-methyltransferase MmcR | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, MmcR, ... | | Authors: | Singh, S, Chang, A, Bingman, C.A, Phillips Jr, G.N, Thorson, J.S. | | Deposit date: | 2009-04-01 | | Release date: | 2010-04-07 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structural characterization of the mitomycin 7-O-methyltransferase.
Proteins, 79, 2011
|
|
3H0W
 
 | | Human AdoMetDC with 5'-Deoxy-5'-[(N-dimethyl)amino]-8-methyl-adenosine | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-deoxy-5'-(dimethylamino)-8-methyladenosine, PYRUVIC ACID, ... | | Authors: | Bale, S, Brooks, W.H, Hanes, J.W, Mahesan, A.M, Guida, W.C, Ealick, S.E. | | Deposit date: | 2009-04-10 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Role of the sulfonium center in determining the ligand specificity of human s-adenosylmethionine decarboxylase.
Biochemistry, 48, 2009
|
|
3RA3
 
 | | Crystal structure of a section of a de novo design gigaDalton protein fibre | | Descriptor: | SODIUM ION, p1c, p2f | | Authors: | Zaccai, N.R, Sharp, T.H, Bruning, M, Woolfson, D.N, Brady, R.L. | | Deposit date: | 2011-03-27 | | Release date: | 2012-08-08 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Cryo-transmission electron microscopy structure of a gigadalton peptide fiber of de novo design
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3GY1
 
 | | CRYSTAL STRUCTURE OF putative mandelate racemase/muconate lactonizing protein from Clostridium beijerinckii NCIMB 8052 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative mandelate racemase/muconate lactonizing protein from Clostridium
beijerinckii NCIMB 8052
To be Published
|
|
3RD0
 
 | | Horse spleen apo-ferritin with bound thiopental | | Descriptor: | 5-ethyl-5-[(2R)-pentan-2-yl]-2-thioxodihydropyrimidine-4,6(1H,5H)-dione, CADMIUM ION, Ferritin light chain, ... | | Authors: | Oakley, S.H, Vedula, L.S, Xi, J, Liu, R, Eckenhoff, R.G, Loll, P.J. | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High resolution view of barbiturate recognition by a protein binding site
to be published
|
|
3H4Q
 
 | |
3H4W
 
 | |
3RDW
 
 | | Putative arsenate reductase from Yersinia pestis | | Descriptor: | Putative arsenate reductase, SULFATE ION | | Authors: | Osipiuk, J, Maltseva, N, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-04-01 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Putative arsenate reductase from Yersinia pestis.
To be Published
|
|
3GJX
 
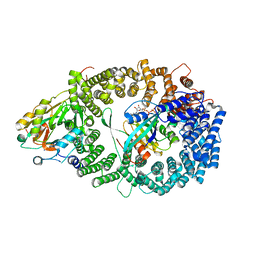 | | Crystal Structure of the Nuclear Export Complex CRM1-Snurportin1-RanGTP | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Monecke, T, Guettler, T, Neumann, P, Dickmanns, A, Goerlich, D, Ficner, R. | | Deposit date: | 2009-03-09 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Nuclear Export Receptor CRM1 in Complex with Snurportin1 and RanGTP.
Science, 2009
|
|
3RCY
 
 | | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing enzyme-like protein from Roseovarius sp. TM1035 | | Descriptor: | GLYCEROL, MAGNESIUM ION, Mandelate racemase/muconate lactonizing enzyme-like protein, ... | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-03-31 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.994 Å) | | Cite: | CRYSTAL STRUCTURE OF Mandelate racemase/muconate lactonizing enzyme-like protein from Roseovarius sp. TM1035
To be Published
|
|
3GMU
 
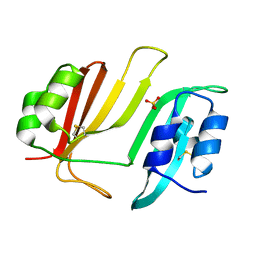 | | Crystal Structure of Beta-Lactamse Inhibitory Protein (BLIP) in Apo Form | | Descriptor: | AMMONIUM ION, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Strynadka, N.C.J, Gretes, M, James, M.N.G. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
3GMX
 
 | |
3IW8
 
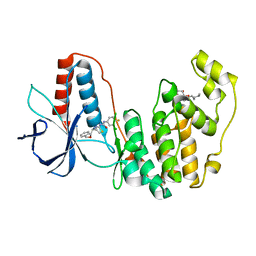 | | Structure of Inactive Human p38 MAP Kinase in Complex with a Thiazole-Urea | | Descriptor: | 1-{4-[(1S)-1-amino-2-(benzyloxy)ethyl]-1,3-thiazol-2-yl}-3-(3-chloro-4-fluorophenyl)urea, Mitogen-activated protein kinase 14, octyl beta-D-glucopyranoside | | Authors: | Gruetter, C, Simard, J.R, Rauh, D. | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-Throughput Screening To Identify Inhibitors Which Stabilize Inactive Kinase Conformations in p38alpha
J.Am.Chem.Soc., 131, 2009
|
|
3RG4
 
 | |
3IHO
 
 | | The C-terminal glycosylase domain of human MBD4 | | Descriptor: | Methyl-CpG-binding domain protein 4 | | Authors: | Amaya, M.F, Xu, C, Bian, C.B, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The C-terminal glycosylase domain of human MBD4
To be Published
|
|
3RGF
 
 | | Crystal Structure of human CDK8/CycC | | Descriptor: | 1,2-ETHANEDIOL, 4-{4-[({[4-CHLORO-3-(TRIFLUOROMETHYL)PHENYL]AMINO}CARBONYL)AMINO]PHENOXY}-N-METHYLPYRIDINE-2-CARBOXAMIDE, Cyclin-C, ... | | Authors: | Schneider, E.V, Boettcher, J, Blaesse, M, Huber, R, Maskos, K. | | Deposit date: | 2011-04-08 | | Release date: | 2011-08-10 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Structure of CDK8/CycC Implicates Specificity in the CDK/Cyclin Family and Reveals Interaction with a Deep Pocket Binder.
J.Mol.Biol., 412, 2011
|
|
3IHT
 
 | |
3RGR
 
 | |
3RHH
 
 | | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-dependent glyceraldehyde-3-phosphate dehydrogenase, SULFATE ION | | Authors: | Malashkevich, V.N, Toro, R, Seidel, R, Garrett, S, Foti, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-04-11 | | Release date: | 2011-05-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of NADP-dependent glyceraldehyde-3-phosphate dehydrogenase from Bacillus halodurans C-125 complexed with NADP
To be Published
|
|
3IJJ
 
 | | Ternary Complex of Macrophage Migration Inhibitory Factor (MIF) Bound Both to 4-hydroxyphenylpyruvate and to the Allosteric Inhibitor AV1013 (R-stereoisomer) | | Descriptor: | (2E)-2-hydroxy-3-(4-hydroxyphenyl)prop-2-enoic acid, (2R)-2-amino-1-[2-(1-methylethyl)pyrazolo[1,5-a]pyridin-3-yl]propan-1-one, 3-(4-HYDROXY-PHENYL)PYRUVIC ACID, ... | | Authors: | Crichlow, G.V, Cho, Y, Lolis, E.J. | | Deposit date: | 2009-08-04 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Allosteric inhibition of macrophage migration inhibitory factor revealed by ibudilast.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
